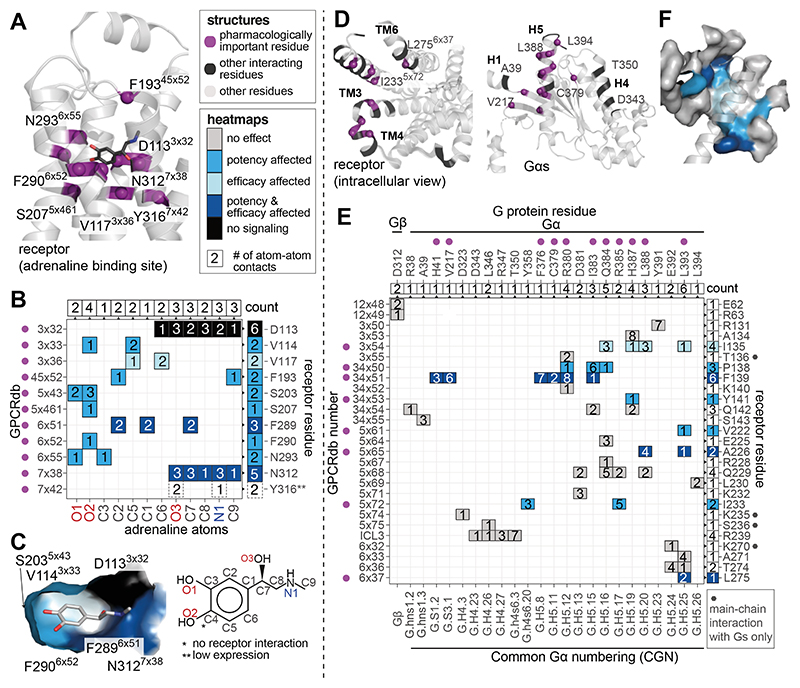
Fig. 3. Effect of mutations in the ligand- and G protein-binding sites.
(A) View of the ligand-binding site, positions are labeled with residue number and GPCRdb number in superscript. Pharmacologically important residues (according to cut-off values, see Methods) are indicated in violet. Not labeled for clarity: V1143x33, S2035x43, F2896x51. (B) Receptor residue-ligand atom contact plot with the ligand atoms on the x-axis and the receptor’s GPCRdb and residue numbers on the y-axis. The number of non-covalent contacts between a receptor residue and ligand atom is shown in each square of the heatmap. The chemical structure of adrenaline below the heatmap indicates the labeling of adrenaline atoms used for the x-axis. Box colors in the heatmap refer to the pharmacological effect of the mutation (efficacy affected (light blue), potency affected (ocean blue), both efficacy and potency affected (dark blue), no measurable signaling (black)). The number of receptor residues contacted by each ligand atom and ligand atoms contacted by each receptor residue are indicated in boxes at the top and right-hand side of the heatmap, respectively. (C) Surface view of the ligand-binding site, where the surface of each receptor residue is colored by the effect its mutation had on signaling. (D) Views of the G protein-binding site on the receptor and the Gs protein. Positions are labeled with residue number and, with GPCRdb numbers. Pharmacologically important residues are indicated in violet and other interacting residues are marked in black. (E) Receptor-G protein residue contact plot based on the structure of β2AR in complex with heterotrimeric Gs (PDB 3SN6). G protein residues are shown on the x-axis and the receptor residues on the y-axis. The receptor residues that contact the G protein through a receptor main-chain contact only are marked with a dark grey hexagon on the right side of the matrix. The number of non-covalent contacts between a receptor residue and a G protein residue are mentioned in each square of the heatmap. See legend for panel B for box colour description. The CGN number (Common G protein numbering scheme) is provided on the x-axis. The number of contacting residues by the receptor and the G protein are shown on the top and right-hand side of the heat map. Residue T68 is not included as it is a contact that is observed only in the structure of β2AR with Gs peptide (PDB 6E67). (F) Surface view of the G protein binding site coloured by mutational effect with helix 5 shown as cartoon.