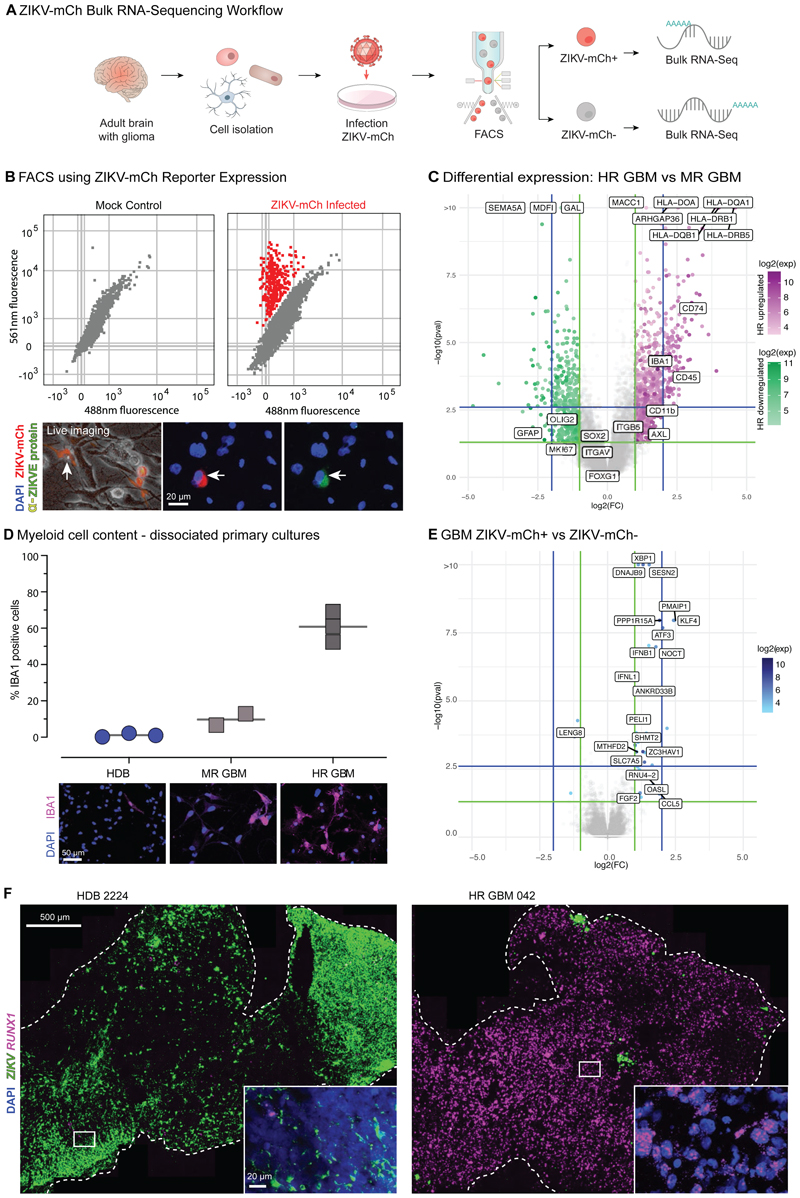
Figure 3. RNA-Sequencing identifies an innate immune signature in ZIKV-refractory GBM.
(A) Experimental scheme: dissociated primary HDB and GBM cultures were infected with ZIKV-mCh reporter virus, then sorted by FACS into mCherry negative and positive fractions for sequencing as shown.
(B) ZIKV-mCherry reporter (ZIKV-mCh) coincides with flavivirus envelope protein expression (α-ZIKVE protein Sigma MAB10216), marking the ZIKV infected fraction.
(C) edgeR differential expression analysis of bulk RNA-Seq in HR GBM (n=3 Cherry paired libraries) vs MR GBM (n=4 Cherry positive/negative paired libraries). Key glioma cell and myeloid cell signature genes are highlighted. HLA-Class II antigen-presenting genes and myeloid cell markers are highlighted.
(D) Cell counts and representative IF images for myeloid cell marker IBA1 in dissociated cell cultures (n=3 each).
(E) edgeR differential expression ZIKV-mCh+ vs ZIKV-mCh- primary GBM culture fractions.
(F) ZIKV-infected HDB and GBM slice cultures: smFISH for ZIKV and myeloid cell marker RUNX1.