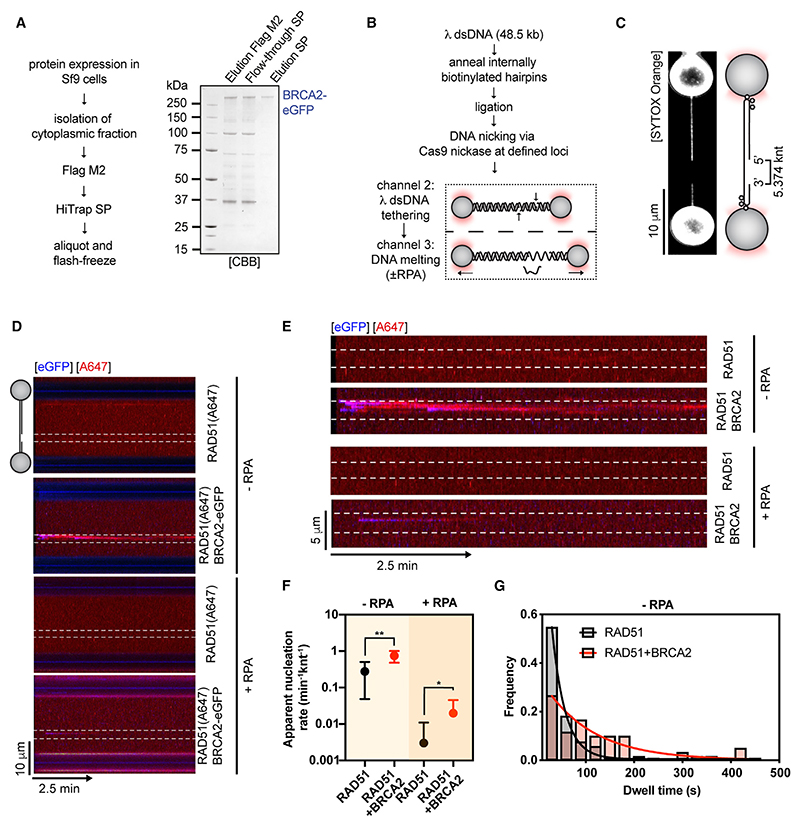
Figure 1. Single-molecule imaging of RAD51 nucleation by full-length human BRCA2.
(A) Purification of full-length human BRCA2-eGFP.
(B) Schematic of the protocol used to generate gapped λ DNA (gDNA) substrates.
(C) Representative image of an asymmetrically positioned ssDNA gap within λ DNA held at 10 pN force.
(D) Kymograph showing the binding of 25 nM RAD51(A647) (red) to gDNA in the presence/absence of 5 nM BRCA2-eGFP (blue) and/or 1.25 nM RPA in the presence of 5 nM SYTOX Orange, 100 mM NaCl, 2 mM MgCl2, 1 mM CaCl2, and 2 mM ATP at ~5 pN force. Position of the ssDNA gap is indicated by dashed lines.
(E) Zoom in on the ssDNA gap region from kymographs in (D).
(F) Quantification of apparent RAD51(A647) nucleation rates in the indicated conditions (n = 5–21 molecules). Dots represent mean. Error bars represent SEM. p values by Student’s t test. n.s., p > 0.05; *p ≤ 0.05; **p ≤ 0.01.
(G) Histograms of dwell times of RAD-51(A647) in the absence (N = 104 clusters) or presence of BRCA2-eGFP (N = 62 clusters). Lines represent exponential fits. See also Figure S1.