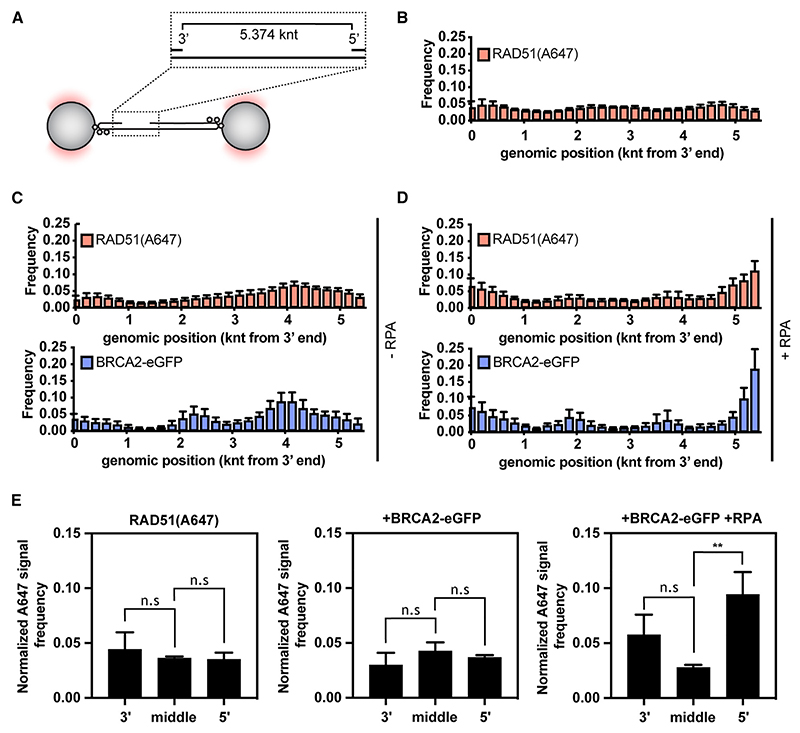
Figure 3. BRCA2 nucleates RAD51 at the edges of ssDNA gap in the presence of RPA.
(A) Schematic of 5.374 knt-long ssDNA embedded within λ dsDNA.
(B) Positional analysis of RAD51(A647) (red) binding along the length of 5.374 knt ssDNA gap. 200 nt bins. n = 5 molecules. Error bars represent SEM. 200 nt bins.
(C) Position analysis of RAD51(A647) (red) or BRCA2-eGFP (blue) binding along the length of 5.374 knt ssDNA gap in the absence of RPA. n = 6 molecules. Error bars represent SEM. 200 nt bins.
(D) Position analysis of RAD51(A647) (red) or BRCA2-eGFP (blue) binding along the length of 5.374 knt ssDNA gap in the presence of 1.25 nM RPA. n = 11 molecules. Error bars represent SEM. 200 nt bins.
(E) Per-bin normalized A647 signal frequency for 3’ and 5’ ds-ssDNA junction (3 and 3 bins) or middle of the ssDNA gap (21 bins) in the indicated conditions. Error bars represent SEM. p values by Student’s t test. n.s., p > 0.05; *p ≤ 0.05; **p ≤ 0.01. See also Figure S2.