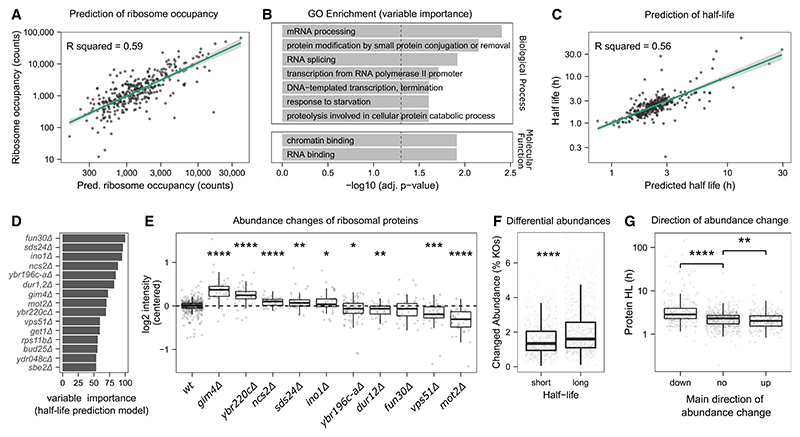
Figure 4. The interdependency of differential protein expression with translation rate and turnover.
(A) Ribosomal occupancies are predicted with an elastic net model. The model was trained on 80% of the proteins (n = 1,392) and applied on the remaining 20% of the proteins (test set, n = 346). The plot shows only proteins from the test set. Ribosomal occupancies were taken from a reference dataset56 and log10-transformed. The proteome data were log2 transformed, centered, and scaled.
(B) Gene Ontology (GO) slim term37 enrichment analysis of the top features selected by the model using a Fisher’s exact test (STAR Methods).
(C) Half-lives are predicted with an elastic net model. The model was trained on 80% of the proteins (n = 1,398) and applied on the remaining 20% of the proteins (test set, n = 348). The plot only shows proteins from the test set. Half-lives were taken from a reference dataset57 and log10 transformed. The proteome data were log2 transformed, centered, and scaled.
(D) The 15 most important KO strains in the regression model for half-lives. The KO strains are ranked by importance and scaled to have a maximum value of 100.
(E) Abundance of ribosomal 60S subunit proteins in 10 KO strains that were selected as the most important feature for the prediction of protein half-life. Protein intensities are centered and log2-transformed. Significance for the comparison to the WT abundance levels (two-sided t test) is shown with asterisks (****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; nsp > 0.05).
(F) Differential abundance of proteins with short (below median) and long (above median) half-lives (****p ≤ 0.0001, Wilcoxon signed-rank test).
(G) Half-lives (in h, log10 transformed) are shown as boxplots for proteins that are predominantly decreased in abundance, increased in abundance, or change in both directions across the KO strains. Directionality was defined as ratios of increased and decreased abundance changes being >75% and <25% quantile for down and up, respectively. Significance (two-sided Wilcoxon signed-rank test with “no direction” as a reference) is shown with asterisks (****p value ≤ 0.0001; **p value ≤ 0.01).
See also Figure S4.