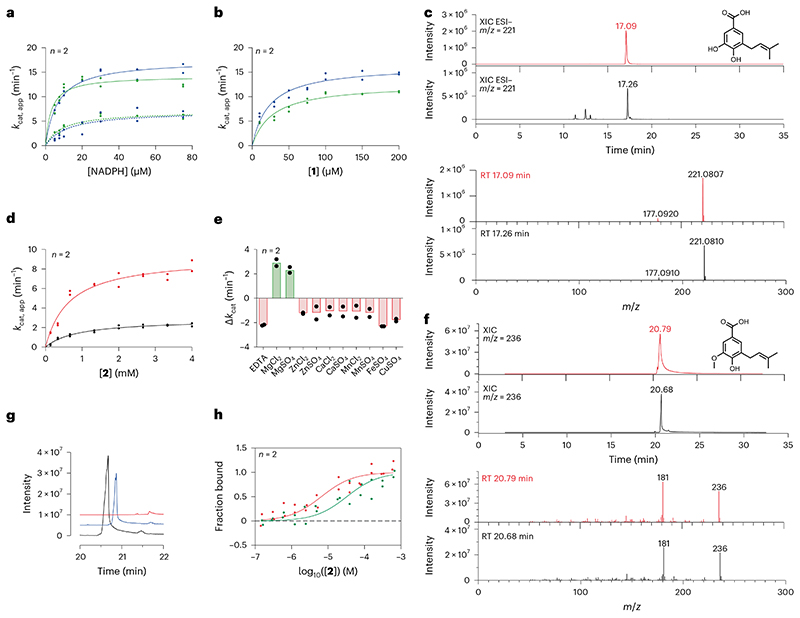
Fig. 2 |. C5 decoration is catalysed by COQ6 and COQ3.
a, Michaelis−Menten kinetics of the FDXR−FDX2 (dashed lines) and FDXR−FDX2−COQ6 (continuous lines) systems in presence of an excess of 1 and increasing NADPH concentrations. Rates were measured with both NADPH (green; Supplementary Fig. 4) and dioxygen (blue) consumption assays. b, Michaelis−Menten kinetics of the FDXR−FDX2−COQ6 system in the presence of saturating NADPH and increasing concentrations of 1. c, UHPLC/HRMS analysis of the overnight COQ6 conversion of 1 to 2. The XICs and ESI−full-scan mass spectra of 2 recorded after the injection of 500 ppm analytical standard and the reaction mixture are shown in red and black, respectively. The theoretical mass of the [M − H]- molecular ion of 2 is 221.0808 Da. It was detected with an error of −0.45 and 0.90 ppm in the analyses of the standard and sample, respectively. d, Michaelis−Menten kinetics of COQ3 in the presence of an excess of SAM with 2 as varying substrate. Activity was measured in the presence (red) and absence of 150 μM MgCl2 (black). e, Bar graph showing the effect of 1 mM EDTA or 150 μM of divalent cations on COQ3 activity. The bars of the histogram show the mean value of independent replicates. f, GC/MS analysis of the overnight COQ3 conversion of 2 to 3. The XICs and full-scan mass spectra of 3 recorded after the injection of 500 ppm analytical standard and of the reaction mixture are shown in red and black, respectively. g, Waterfall plot showing a qualitative analysis of the GC peak of 3 generated by COQ3 in the presence (black) or absence (blue−time offset 0.2 min) of 150 μM MgCl2 and in presence of 1mM EDTA (red−time offset 0.4 min). h, Microscale thermophoresis binding affinity curves of 2 binding by NHS-labelled COQ3 in presence (red) and absence (green) of 150 μM MgCl2. Individual data points corresponding to n = 2 independent measurements are shown in a, b, d, e and h. RT, retention time.