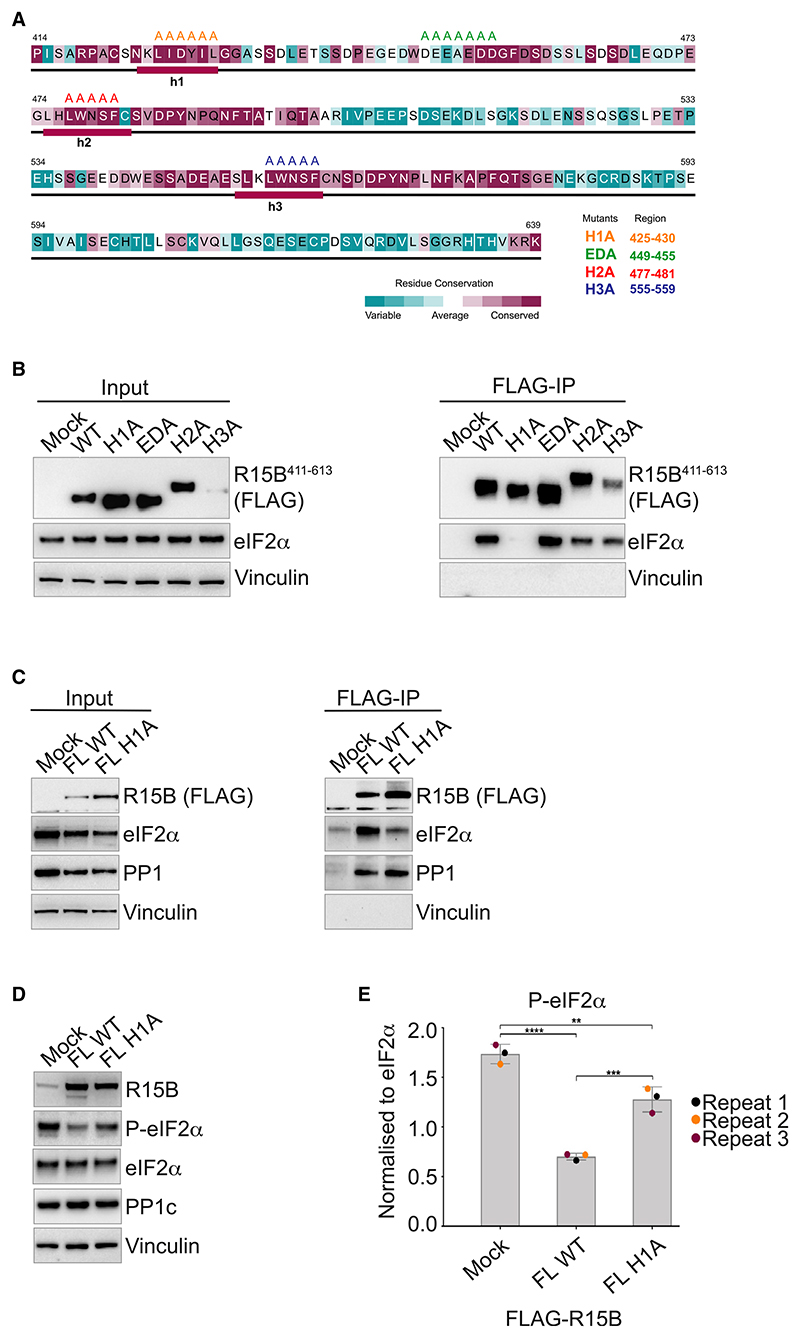
Figure 5. Identification of a mutant of R15B defective in substrate binding.
(A) Sequence conservation of R15B414–639. The residues are colored according to ConSurf41 conservation scores from cyan (variable) to burgundy (conserved). The consensus secondary structure, predicted using Jpred42 and PsiPred,36 is shown below the corresponding sequence. The secondary structural elements are denoted as follows: rectangle, helix; line, random coil. The substitutions of the residues targeted for mutagenesis are shown on the top with different mutants shown in different colors.
(B and C) R15B411–613 (B) or full-length (C) wild-type (WT) or mutants were transfected into HEK 293T cells (input) and immunoprecipitated using anti-FLAG M2 magnetic beads (FLAG-IP). Immunoprecipitated complexes were eluted and analyzed on a 4%–12% bis Tris Plus gel. Proteins were detected by immunoblotting with FLAG, eIF2α, PP1, and vinculin antibodies. Representative results of at least 3 experiments are shown.
(D) Activity of transfected R15B full-length WT or H1A assessed by decreased levels of P-eIF2α. Proteins were detected by immunoblotting with R15B (R15B-4D11), P-eIF2α, eIF2α, PP1, and vinculin antibodies. Representative results of at least 3 experiments are shown.
(E) Quantifications of P-eIF2α from 3 experiments such as the one shown in (D). Data are mean ± SD. (n = 3). **p < 0.01, ***p < 0.001, ****p < 0.0001, as determined by one-way ANOVA.