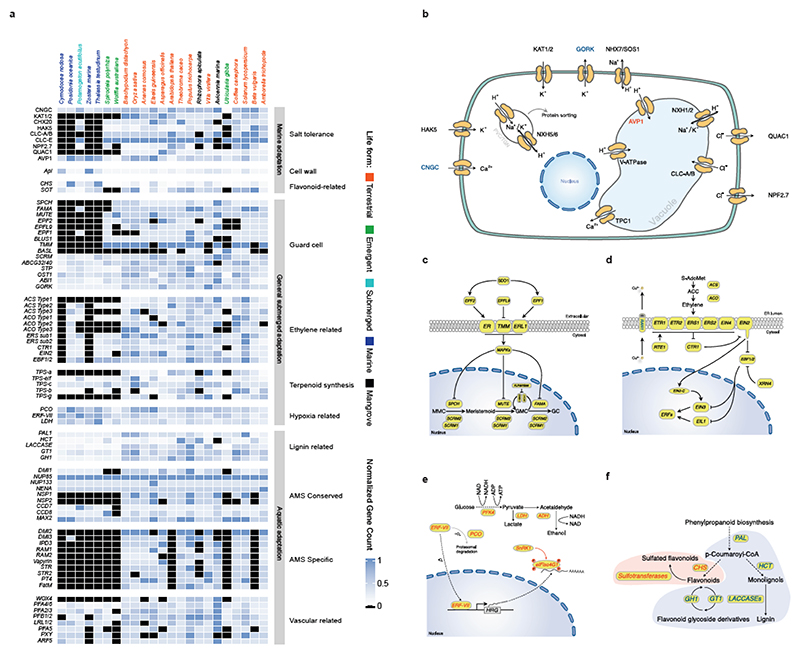
Figure 3. The loss, contraction, and expansion of gene families involved in the adaption to a marine environment.
a) The normalized gene copy numbers for 4 seagrasses and 19 representative non-seagrass species. The normalization on the family dataset divides the gene count number of each species by the largest gene copy number within that family. The species order on the top of the heatmap is the same as that in Figure 1. The colors correspond to the different life-forms. The orange ones are terrestrial species; the green ones are emergent species (floating-leaved); the light blue ones are submerged species; the navy-blue ones are marine species (seagrasses) and the black ones are mangroves b) Salt stress signaling implies different ion channels. HAK5 encodes HIGH-AFFINITY POTASSIUM TRANSPORTER 5; CNGC, CYCLIC NUCLEOTIDE GATE CATION CHANNELs; AVP1 encodes Vacuolar H+-PPases c) Stomata differentiation from meristemoid mother cells (MMC) to guard mother cell (GMC), to guard cells. d) Ethylene synthesis and signaling. e) The hypoxia-responsive signaling in which the direct (ERF-VII) and indirect responsive (SnRK1) pathways are expanded. The rate-limiting enzyme (encoded by PFK4) in the glycolysis pathway, along with Lactate dehydrogenase (encoded by LDH), a rate-limiting enzyme in fermentation, are also expanded. F) Simplified schematic of the lignin and flavonoid biosynthesis pathways. Only steps that have significantly changed are shown. PAL encodes phenylalanine ammonialyase, which is the gateway enzyme of the general phenylpropanoid pathway; CHS encodes chalcone synthase, which is the first enzyme of flavonoid biosynthesis that directs the metabolic flux to flavonoid biosynthesis; GT1 encode flavonoid glycosyltransferases, which catalyze the final step of flavonoid biosynthesis to generate various flavonoid glycoside derivatives; GH1 encode flavonoid beta-glucosidase & myrosinase, which are responsible for the recycling of carbohydrate-based flavonoids; HCT encode Hydroxycinnamoyl‐CoA shikimate/quinate hydroxycinnamoyl transferase, channels phenylpropanoids via the “esters” pathway to monolignols”; LACCASEs encode the final enzymes in the pathway that oxidize monolignols to facilitate their polymerization into lignin. Panels d) e) f) genes in red are expanded; blue means contracted; The dashed line in the pathway means multiple metabolic steps.