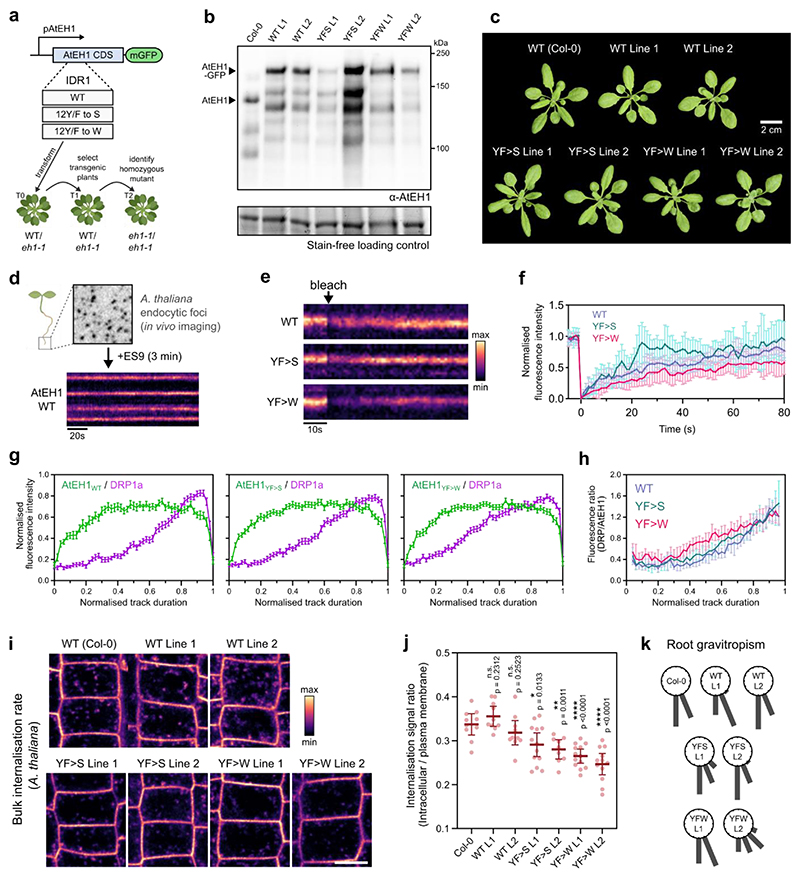
Extended Data Fig. 7 (Related to Figure 7). pAtEH1:AtEH1-GFP reporter validation and functionality.
a, Schematic of the generation of pAtEH1:AtEH1-GFP rescue lines. b, Western blot of A. thaliana protein extracts using an antibody directed against AtEH1, indicating absence of endogenous AtEH1 in the recued lines. Lower molecular weight bands are degradation products. Strain free gel is shown as a loading control. c, Representative plant rosette image of WT Col-0 and AtEH1 rescued lines. d-f, Inhibition of endocytosis in pAtEH1:AtEH1-GFP seedlings using the chemical inhibitor ES9 (3 min, 10 μM). Kymograph shows immobile endocytic foci with stable fluorescence intensity over time (d). Kymograph (e) and fluorescence intensity plot (f) from FRAP experiments of individual endocytic foci from pAtEH1:AtEH1WT-GFP, pAtEH1:AtEH1YF>S-GFP, and pAtEH1:AtEH1YF>W-GFP seedlings. Plotted values indicate mean ± SD. Half recovery times (t1/2) are 22.08s, 11.95s, 28.81s, respectively. g, Full intensity profiles from kymograph analysis of endocytic events from root cells of A. thaliana AtEH1:AtEH1-GFP/eh1-1 lines in a DRP1a-mRFP/drp1a background (see also Fig. 7g,h). Plotted values indicate mean ± SEM. h, Fluorescence intensity ratio of DRP1a compared to AtEH1 from data in (g). Plotted values indicate median ± 95% CI. (i,j) Endocytic flux experiment. Seedlings were treated with the lipophilic dye FM4-64 (2μM, 10min) and root cells were imaged (i). j, Quantification of FM4-64 internalisation. Bars indicate mean ± 95% CI. Statistics indicate significance to WT; n.s. not significant, * p<0.05, ** p<0.01, **** p<0.0001, unpaired t-test. k, Gravitropism assay. Roots grown on ½ MS agar plates were gravistimulated by turning them 90°. The gravitropism plot indicates the angle of the root tip in 22.5° bins. n= 20 roots for each genotype. Scale bar = 10 μm (d). Data in i-j represents pooled data from three independent repeats. The experiment in b, f and k was performed two, two and three times, respectively with similar results. Sample sizes (f, j, k) are indicated in Source Data. Source data are provided as a Source Data file.