Abstract
RHO GTPases have been traditionally associated with protumorigenic functions. While this paradigm is still valid in many cases, recent data have unexpectedly revealed that RHO proteins can also play tumor suppressor roles. RHO signaling elements can also promote both pro- and antitumorigenic effects using GTPase-independent mechanisms, thus giving an extra layer of complexity to the role of these proteins in cancer. Consistent with these variegated roles, both gain- and loss-of-function mutations in RHO pathway genes have been found in cancer patients. Collectively, these observations challenge long-held functional archetypes for RHO proteins in both normal and cancer cells. In this review, I will summarize these data and discuss new questions arising from them such as the functional and clinical relevance of the mutations found in patients, the mechanistic orchestration of those antagonistic functions in tumors, and the pros and cons that these results represent for the development of RHO-based anticancer drugs.
Introduction
The 18 RHO GTPases present in humans can be classified according to both structural and homology criteria in the CDC42 (CDC42, RHOQ, and RHOJ), RAC (RAC1, RAC2, RAC3, and RHOG), RHOA (RHOA, RHOB, and RHOC), RHOD (RHOD and RHOF), RHOH, RHOU (RHOU and RHOV), and RND (RND1, RND2, and RND3) subfamilies [1]. Some studies also include among those proteins the members of the RhoBTB and Miro families, although these ‘atypical’ GTPases are highly divergent from the rest of Rho proteins in terms of structure, overall amino acid homology, subcellular localization, and biological functions [1]. Rho proteins were initially linked to the regulation of cytoskeletal structures, cell shape, cell migration, and cell polarity [2]. Later on, they were associated with the regulation of cell cycle progression, cell survival, and cell type-specific responses such as immune responses, angiogenesis, vascular reactivity, and neurogenesis [1,3–7]. Given this central role in cell signaling, the deregulation of these pathways is also linked to the development of a large variety of diseases [2,4,8,9]. In particular, the action of RHO GTPases has been historically associated with the acquisition of malignant features by cancer cells [10–13]. While this association was initially established using cancer cell models in plastico, more recent information obtained from both mouse models and high-throughput genomics techniques has given further impetus to this idea. However, these new data have also unveiled the paradoxical association of some of these proteins with tumor suppression mechanisms. Adding further complexity to the variegated role of these proteins in cancer, it is now known that many RHO signaling elements can contribute to tumorigenesis using noncanonical, GTPase-independent mechanisms. These observations highlight the key role of RHO proteins in cancer but, at the same time, challenge widely established functional archetypes in the field and the therapeutic feasibility of these pathways. In this review, I will present recent advances in the understanding of RHO-regulated pathways in cancer, highlight some of the shadows existing in the field, and the challenges we still face to develop new RHO-based therapies. Given the scant information available for the less conventional RHO family members, I will focus my attention on the classical CDC42, RAC, and RHOA subfamilies.
The RHO GTPase cycle
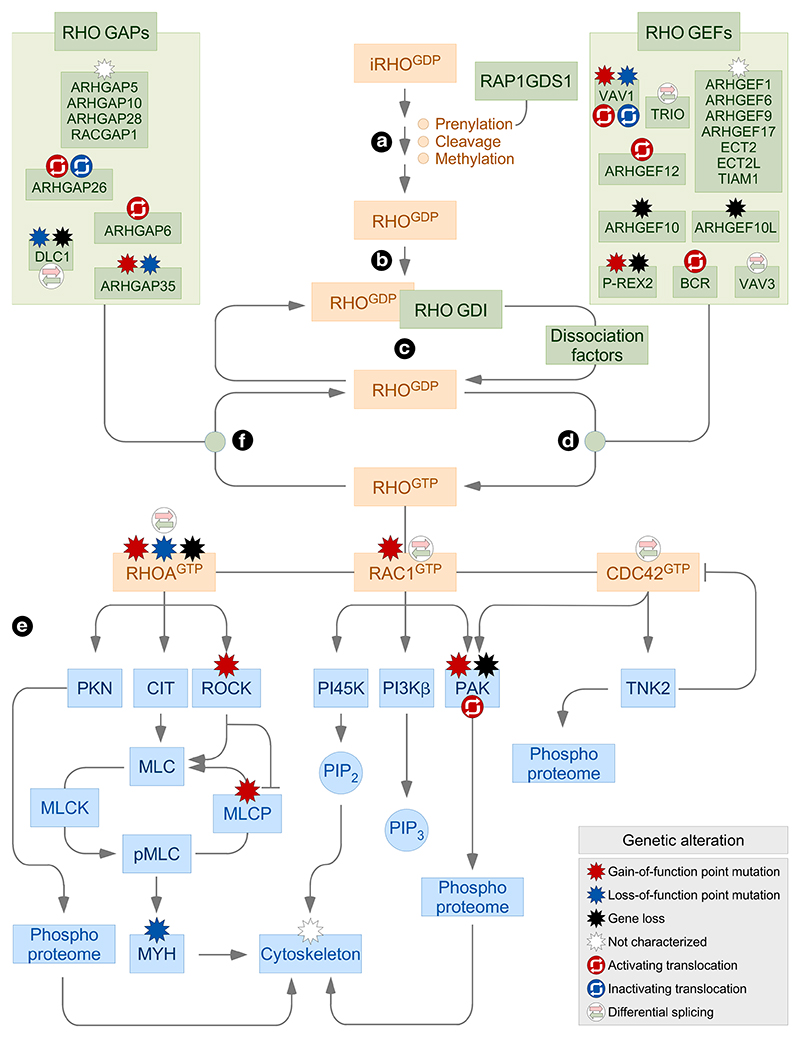
RHO GTPases are subjected to multiple regulatory layers affecting their subcellular localization, intracellular dynamics, signaling state, and final downstream signaling output [1,14] (Figure 1). As most RAS superfamily proteins, the maturation of the freshly translated RHO GTPases entails a stepwise mechanism that includes the incorporation in the cytosol of a geranylgeranyl group onto the cysteine residue located on the so-called C-terminal CAAX box, the translocation of the GTPases from the cytosol to the endoplasmic reticulum, the subsequent cleavage of the C-terminal AAX tripeptide of the GTPases, and the ensuing methylation of the α-carboxyl group of the newly exposed C-terminal isoprenylcysteine residue (Figure 1, point a). There are variations in this maturation process in some cases, such as the incorporation of a farnesyl rather than a geranylgeranyl group (e.g. RHOB) or the addition of palmitate groups onto cysteine residues located outside the GTPase CAAX box (e.g. RAC1 and RHOB) [1,14]. Some of these modifications are not trivial from a functional point of view. For example, the farnesylated and geranylgeranylated states are associated with the tumor promotion and suppression functions of RHOB, respectively [15]. The alternative prenylation involves the participation of additional regulatory molecules, as is the case of different isoforms of the Rap1 GDP dissociation stimulator (RAP1GDS, also known as small GTPase GDS) [1,14,16].
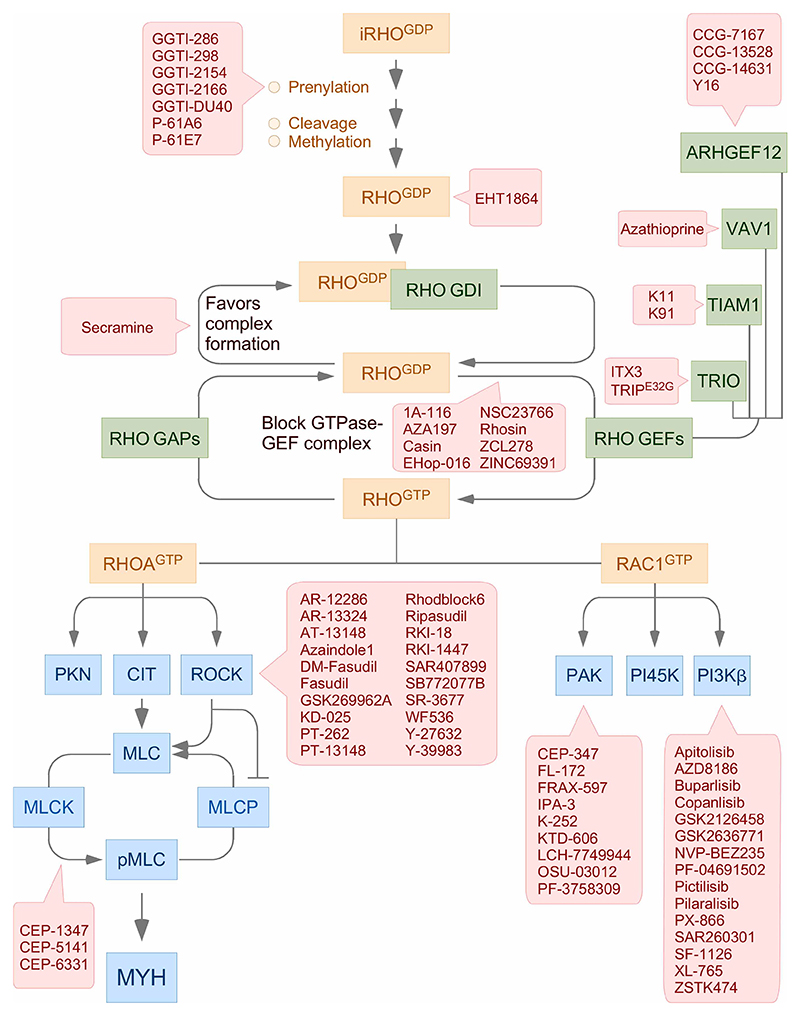
Figure 1. Main regulatory cycle of RHO proteins.
GTPases, regulators, and effectors are shown in brown, green, and blue boxes, respectively. Genetic alterations found in some of these signaling elements in tumors are shown (inset). Inactivation steps are shown with blunted lanes. The indicated mutations have been collected from information present in the cBioPortal (http://www.cbioportal.org), St. Jude Cloud PeCan (https://pecan.stjude.cloud/home), and recent publications on this topic. iRHO, immature RHO; RHOGDP, GDP-bound RHO; RHOGTP, GTP-bound RHO; CIT, citron kinase; PI45K, phosphatidylinositol-4-phosphate 5-kinase; MLC, myosin light chain; MLCK, myosin light chain kinase; MLCP, myosin light chain phosphatase; MYH, myosin heavy chain; PIP2, phosphatidylinositol (4,5) biphosphate; PIP3, phosphatidylinositol (3,4,5) triphosphate.
The fully processed GTPases are maintained locked in an inactive GDP-bound state through the formation of stoichiometric complexes with RHO GDP dissociation inhibitors (GDIs) in the cytosol (Figure 1, point b). These proteins contribute to that process by blocking the spontaneous release of GDP from the GTPases and, in addition, by sequestering the prenyl group of the GTPases inside a hydrophobic cavity present in the GDI 3D structure. They also play positive roles in the activation of RHO proteins, since they can protect them from proteolytic degradation and, upon cell stimulation, dock the GTPases in specific membrane subregions [1,14]. Most RHO proteins have to undergo the exchange of GDP by GTP molecules during signal transduction to acquire an active state compatible with downstream effector binding. This process requires two intertwined steps. The first one involves the release of the GTPases from the RHO GDI complexes (Figure 1, point c), a process mediated by the phosphorylation of RHO GDIs by SRC, protein kinase C, and members of the p21-activated kinase (PAK) family [1,14]. The participation of membrane lipids, RHO GDI-releasing factors, and RHO translocators has been also reported in specific signaling contexts [1,17–19]. The released GTPases then undergo rapid exchange of guanosine nucleotides, a process catalyzed by the enzymes known as RHO GDP/GTP exchange factors (GEFs) (Figure 1, point d). The human genome encodes close to 80 GEFs that are highly variable in terms of catalytic specificity and domain structure. RHO GEFs can be subclassified according to the catalytic domain present in them in the diffuse B-cell lymphoma (Dbl) and dedicator of cytokinesis (Dock) subfamilies. Experimental evidence indicates that this complexity ensures enough flexibility to fine-tune the spatio-temporal activation and signaling output of RHO proteins [1,20]. The exchange of nucleotides promotes a conformational change in the RHO switch I and switch II effector regions that, in turn, makes it possible the interaction of the GTP-bound GTPases with the proximal downstream effectors. To date, ~60 RHO proximal effectors have been identified. Although highly heterogeneous from a structural and functional point of view, most of them can be ascribed to a limited number of functional classes: (a) direct regulators of the F-actin cytoskeleton and cell polarity. (b) A grab bag of proteins that include, for example, regulators of multiprotein enzyme complexes (e.g. NADPH oxidase) and transcriptional factors (STAT3). (c) Serine/threonine kinases belonging to PAK, RHO-associated coiled-coil containing protein kinase (ROCK), and protein kinase N (PKN1) families (Figure 1, point e). (d) Protein tyrosine kinases (TNK2, also known as ACK1) (Figure 1, point e). (e) Phospholipid kinases such as phosphatidylinositol 3-phosphate kinase (PI3K) β and phosphatidylinositol-4-phosphate 5-kinase (Figure 1, point e) [1,21]. The recognition of the downstream effectors depends on structural cues present on the two RHO switch regions and, in some cases, other GTPase moieties. These cues constraint the type of effectors recognized by each RHO subfamily and, in many cases, specific members of the same RHO subfamily. For example, PAKs can bind to RAC1 and CDC42 but not to RAC2, RHOG, or any of the RHOA subfamily members. Likewise, ROCK proteins physically interact with active RHOA subfamily members but not with those belonging to the RAC and CDC42 subgroups (Figure 1, point e) [1]. At the end of the stimulation cycle, RHO GTPases undergo the hydrolysis of the bound GTP molecules to go back to the inactive, GDP-bound state. This step is catalyzed by GTPase-activating proteins (GAPs) (Figure 1, point f). As in the case of RHO GEFs, the GAP family is composed of a large variety of members that are quite different in terms of catalytic, structural, regulatory, and effector properties [1,20].
RHO GTPases can be further regulated by mechanisms outside this basic regulatory cycle, including transcriptional regulation, differential splicing, microRNA-mediated transcript stability, protein steady-state levels, posttranslational modifications, sequestration in endosomes, time of residence in membranes, and F-actin-dependent cytoskeletal events that favor more stable cycles of activation during cell stimulation. Some of these layers are also used to modulate the function of RHO GTPase regulators and effectors, thus ensuring a perfect control of the signaling output from these pathways [1,12,14,18,22].
Protumorigenic functions of RHO GTPase-regulated pathways
According to the canonical view of RHO GTPase pathways in cancer, it has been historically assumed that the RHO GEFs, the GTPases themselves, and many of the downstream effectors play positive roles in tumorigenesis. Conversely, this functional archetype postulates that RHO GAPs must antagonize the foregoing process. Consistent with this view, extensive work using gain-of-function strategies demonstrated that mutant versions of RHO GEFs and, to a lesser extent, RHO proteins were oncogenic using focus formation assays. Conversely, the use of loss-of-function studies showed that many RHO signaling elements were required for either the overall fitness or specific malignant properties of cancer cells. In contrast with these in plastico data, one of the most puzzling observations in the field until this decade was the lack of detection of mutations in genes encoding RHO signaling elements in human tumors. It was reasoned at that time that this was probably due to the fact that RHO pathways required the preservation of normal cycles of activation/deactivation to promote tumorigenesis. However, the extensive sequencing of tumor genomes carried out during this last decade has unveiled the presence of gain-of-function mutations in genes encoding RAC1 [23–38], RAC GEFs (P-REX2 and VAV family) [27,39–46], and RAC downstream elements (PAK1, PAK4, and PAK5) [26,47] in patients (Figures 1 and 2A–C). Gain-of-function mutations in RHOA and ROCK1 have also been found at lower frequencies in some human tumors [28,29,43,48–54] (Figures 1 and 2A–C). Although dispensable for leukemogenesis, the RHOA GEF domain of the BCR protein present in the chimeric p210BCR-ABL oncoprotein seems to play roles in the determination of the leukemia subtype that eventually develops in patients [55]. The mutational and epigenetic silencing of DLC1, a gene encoding a RHOA GAP, is also a frequent event in human tumors [52,56] (Figure 1). Further analyses of cancer genomes have revealed the presence of additional hotspot mutations in other genes encoding RHO signaling elements, although most of them remain to be investigated at the functional level [34,35,57,58] (Figure 1; further data can be analyzed at the inTOgene database using the link: https://www.intogen.org/search). To date, very few cases of gain-of-function mutations have been found in the CDC42 gene [31] (Figures 1 and 2A).
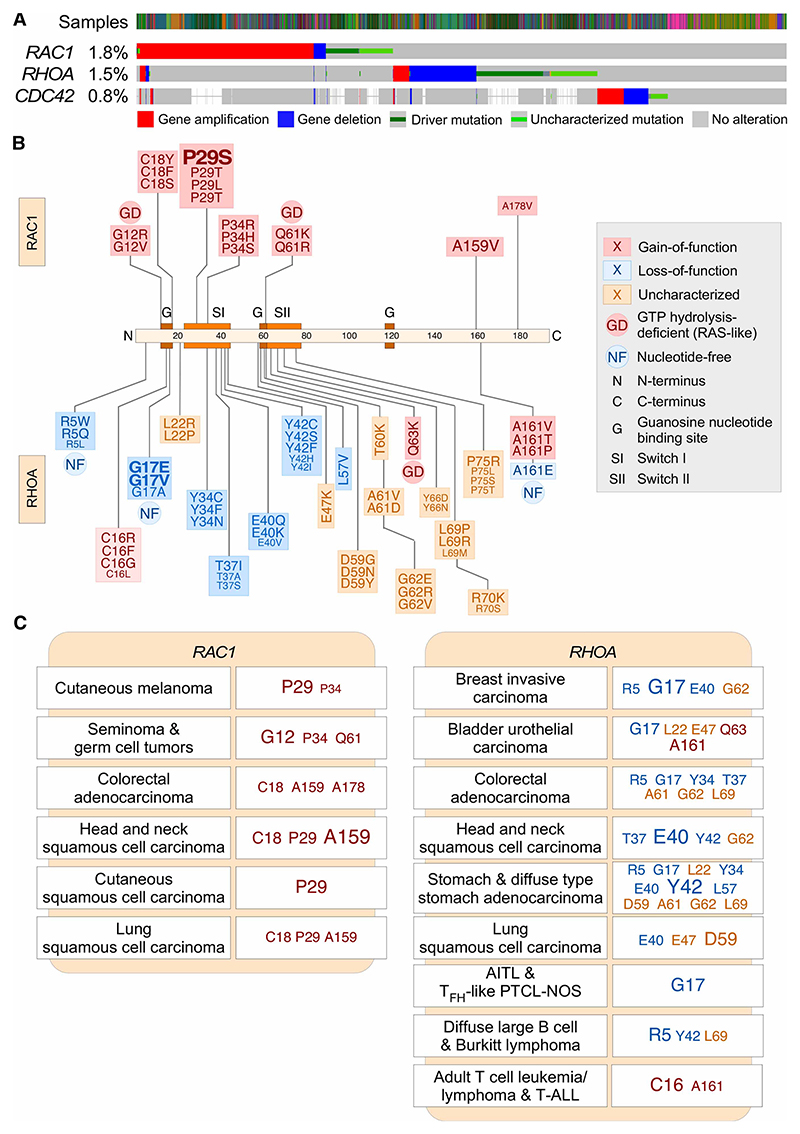
Figure 2. Mutational pattern of RAC1, RHOA, and CDC42 genes in human tumors.
(A) cBioPortal-generated data depicting the genetic alterations found both in human tumors and cancer cell lines for the indicated RHO family genes (left). The type of mutation is indicated at the bottom. The frequency of genetic alterations found for each gene in the total number of samples that have been sequenced is indicated on the left. (B) Depiction of the main mutations found in RAC1 (top) and RHOA (bottom) genes. The most frequently mutated amino acid positions are shown in larger font. See inset for further information. (C) Examples of main mutations found in RAC1 (left) and RHOA (right) genes in the indicated tumors. The most frequent mutations are shown in larger fonts. These data were collected as in Figure 1. AITL, angioimmunoblastic T-cell lymphoma; PTCL-NOS, peripheral T-cell lymphoma-not otherwise specified.
The most frequent gain-of-function mutations found in RHO family genes alter codons that lead to the generation of proteins with high rates of intrinsic GDP/GTP exchange [23,24,43,53,59]. In the case of RAC1, the main targeted residues include Pro29 and, to a lesser extent, the Cys18, Pro34, Ala159, and Ala178 residues (Figure 2B,C). In the case of RHOA, these gain-of-function mutations target the positions Cys16 and Ala161 (Figure 2B,C; note that the amino acid positions in RHOA are shifted +2 positions relative to those found in most RHO and RAS GTPases). However, mutations that disrupt GTP hydrolysis are found enriched in specific tumor subtypes. These mutations mostly target codons similar to those found in classical RAS proteins [60] such as Gly12, Gln61 (both in RAC1), and Gln63 (in the case of RHOA) (Figure 2B,C). This odd mutational spectrum, combined with the relatively low frequency of all these genetic alterations in patients, probably explains the lack of detection of mutations in these genes during the pregenomics period.
Several mechanistic and functional questions arise from the foregoing data. One of them is the reason for the characteristic type of oncogenic mutations found in the RAC1 and RHOA. A possible answer to this issue is that those mutations are selected because they confer a specific advantage to cells when compared with the potentially stronger, GTP hydrolysis-deficient counterparts. Evidence for this type of differential signaling is, in fact, available in the literature [61,62]. If this were the case, the advantage of these weak signaling mutants should be tumor-specific given that the RAC1 Gly12 and Gln61 are the most frequently targeted residues in other tumors types such as seminomas and germ cell tumors (Figure 2C). However, the segregation of subsets of RAC1 mutations in different tumor types suggests that the spectrum of those mutations can be merely reflect the type of carcinogen to which the cells that originated the tumors were originally exposed to. This idea is consistent with the observation that the codon encoding Pro29, the most frequently targeted RAC1 residue in cutaneous melanoma [23,25,26,31] (Figure 2C), exhibits the typical mutational signature associated with UV light exposure [23]. In line with this, the frequency of this mutation directly correlates with the sun exposure habits of the melanoma patient cohorts analyzed [23]. More work should be done in any case to further clarify this issue. A more important question is the role played by these mutations in tumor development. Although answering this question requires further experimentation, current evidence from either mouse or Zebra fish models suggests that the gain-of-function mutations in RAC1, RHOA, RHO GEF-encoding genes (PREX2 and VAV2), PAK1, and ROCK are not fully autonomous to drive the transformation of primary cells, although in some cases hyperplasic, preneoplasic-like phenotypes are observed. However, most of those models do show higher tumor formation rates when combined with other protumorigenic genetic alterations (Figure 3) [1,7,63–66] (in the case of Vav2, unpublished observations from our laboratory). Given the disparity of mutations found in tumors, we cannot rule out the possibility at this moment that some of them could act as oncogenic drivers per se. Addressing this issue will require further studies using ad hoc-designed animal models.
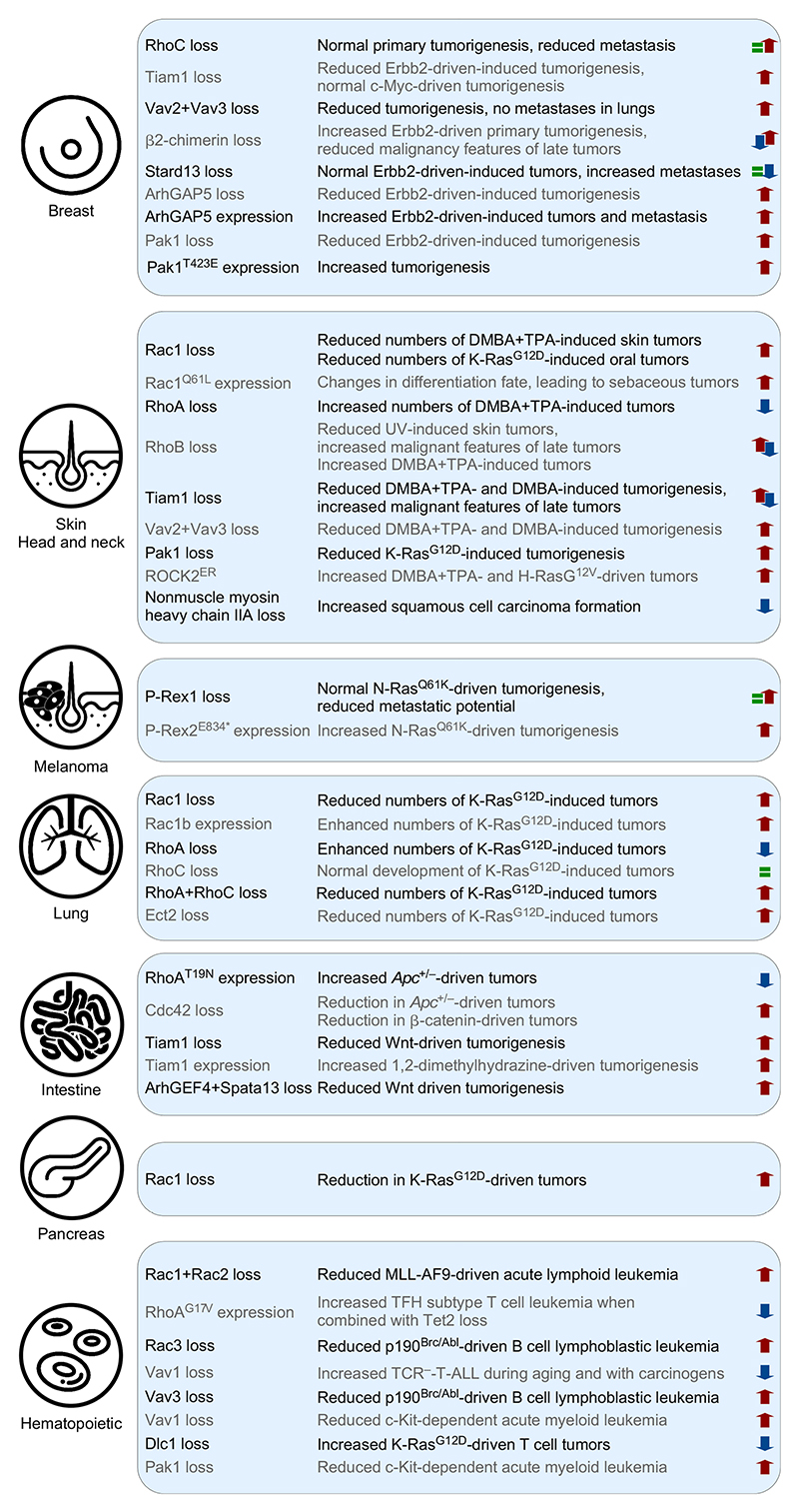
Figure 3. Effects of indicated alterations in RHO signaling elements in tumorigenic processes in mouse models.
Protumorigenic (top-pointing red arrows), antitumorigenic (bottom-pointing blue arrows), and no significant effects (green equal sign) are indicated. ArhGEF4 and Spata13 are also known as Asef1 and Asef2 GEFs, respectively. DMBA, 7,12-dimethylbenz(a) anthracene; TPA, 12-O-tetradecanoylphorbol-13-acetate; APC, adenomatous polyposis coli; MLL-AF9, mixed lineage leukemia-MLLT3 (AF9) fusion oncoprotein; TCR–, T-cell receptor negative.
Regardless of the above data, the analysis of cancer genomes indicates that mutations affecting RHO signaling pathways are found at low frequency in a very limited spectrum of tumor types (Figure 2A). It is unavoidable therefore to address again the question made before the cancer genome sequencing era: why cancer cells do not consistently develop mutations in RHO pathways at high frequencies? The most obvious answer to this question, which most of us will probably dislike, is that the RHO pathways are totally irrelevant for the fitness of most cancer types. Another possibility is that the RHO pathways can be spuriously activated in cancer cells by alternative mechanisms. A strong emphasis has been made, for example, in the frequent detection of cancer-associated gene copy alterations and protein abundance changes in RHO signaling elements as a source of such deregulation [12]. Some of these changes are indeed recurrently found in tumors (Figure 2A) and, in the case of RHO downstream elements such as PAK1, PAK4, ROCK2, and TNK2, have being formally demonstrated that they indeed contribute to the fitness of cancer cells [47,52,54,67]. However, other data related to this type of deregulation should be taken with caution for manifold reasons. Firstly, the changes in the number of copies of a given gene can be merely a passenger event due to the presence of a nearby driver gene. Secondly, the changes in the abundance of a given signaling element might actually elicit functional consequences opposite to the expected ones. For example, the low expression of RHO GDIs can lead to enhanced degradation of RHO proteins rather than to increased availability of free RHO GTPases for signaling as originally surmised. The same applies to the overexpression of RHO GTPases, an event that can be easily buffered in cells through the usual stoichiometric association of GTPases with RHO GDIs. Finally, the increased abundance of a given protein can be a consequence rather than the cause of the malignant feature of cancer cells. This is probably the case, for example, of RHO signaling elements associated with the regulation of cytokinesis (i.e. ECT2 and RACGAP1). Our experience with VAV and RHO proteins also suggests that many immunohistological analyses used to demonstrate protein overexpression on tumor sections are based on the use of poorly validated antibodies both in terms of isoform specificity and, even, on the nature of the epitope that is actually detected on the tissue sections. Other mechanisms that could lead to the spurious activation of RHO pathways include alterations in the multiple regulatory layers that control the steady-state levels and activation dynamics of RHO signaling elements in cancer cells (see above). Many of these alterations have been observed in cancer cells in culture [1,12,14,18,22], although until now there is no experimental evidence indicating that such alterations do have a major effect in either human tumors or animal models. The only exception is perhaps the frequent alteration observed in the splicing of the RAC1 hnRNA in many tumors (Figure 1). This change leads to the expression of a fast cycling isoform of wild-type RAC1 that, according to cell and mouse model data (Figure 3), is clearly associated with the induction of protumorigenic effects in cells [68–73]. Differential spliced forms for CDC42, RHOA, and GEFs (TRIO and VAV3) have also been observed in cancer cells [68,74–78] (Figure 1), suggesting that this type of deregulation might represent a common event favoring the engagement of these pathways in many tumor types. Finally, it is possible that the hyperactivation of the endogenous wild-type RHO pathways via autocrine loops, upstream oncogenes, and signaling cross-talk could be sufficient to provide a selective advantage to cancer cells [12]. Along those lines, the use of knockout and transgenic mice has demonstrated that many endogenous RHO GTPases, regulators, and effectors are indeed required for the primary tumorigenesis and/or metastatic properties of many tumor types [64–66,72,79–107] (Figure 3). Recent genome-wide shRNA- and CRISPR/Cas9-based loss-of-function studies have also made it possible, for the first time, to visualize in a high-throughput basis the implication of all known RHO signaling elements in the viability of a large number of cancer cell lines [108–110]. The readers can access these data in publicly available data portals derived from the Achilles (https://portals.broadinstitute.org/achilles), Drive (https://oncologynibr.shinyapps.io/ drive/), and DepMap (https://depmap.org/rnai/) projects.
Given that most work on RHO GTPases has been narrowly focused on cytoskeletal-related biological processes and canonical pathways using cells in plastico, we still have a very limited information about the pathobiological changes induced by them in tumors. Recent observations suggest that these proteins play more variegated roles than previously anticipated. Those include the regulation of autocrine and paracrine loops important for tumor growth and the remodeling of the tumor microenvironment [100], nucleolar functions linked to either efficient ribogenesis or the elimination of nucleolar stress in cancer cells [102,111], the regulation of both centrosome and chromosome stability [112–114], the regulation of the YAP/TAZ pathway [115], and the engagement of pathways that evade the antitumoral immune responses [116]. Other interesting protumorigenic actions include the PAK-mediated enhancement of the signaling output from the RAS–ERK and RAS–PI3K pathways [47,117,118]. The activity of RAC-dependent pathways is particularly relevant from a clinical point of view since they favor the proliferation of cancer cells, the maintenance of tumor initiating cells, and the development of drug resistance [118–131]. The contribution of RHOA subfamily and ROCK-dependent pathways to cancer cell stemness and drug resistance has been also reported [52,54,120,127,132–134]. Further studies on the biological outputs of these proteins using both animal models and more close-to-the-clinic experimental scenarios will probably expand the panoply of RHO-regulated functions in cancer cells in the coming years.
Tumor suppressor activities of RHO-dependent pathways
The puzzling discovery of both dominant negative and loss-of-function RHOA gene mutations in human tumors underscores the limited understanding that still exists on these pathways [29,30,32,34,38,41,43,45,48,49,53,57,135–146] (unpublished data can be checked at the cBioPortal database: http://www.cbioportal.org/index.do?session_id= 5a98e6f6498eb8b3d5650200) (Figure 2). They also highlight the limitations of using cancer cells in 2D cultures to approach the functions of these proteins, since the large majority of these studies have not unveiled these new roles for the RHOA pathway. These mutations can be classified in three main subclasses (Figure 2B): (a) mutations that generate inactive proteins that cannot bind guanosine nucleotides. These mutant proteins must also trap the upstream GEFs in nonproductive complexes, leading to the inhibition of the stimulation of wild-type RHOA (when the RHOA mutation is in heterozygosis) and the rest of GTPase substrates of those GEFs [147]. These mutations target the RHOA Gly17 residue and, to a lesser extent, other positions of the GTPase (Arg5, Gly14, Thr19, and Leu57). (b) Mutations targeting residues on the RHOA switch I region that are critical for the association with the downstream elements such as PKN (Tyr37 and Tyr42) and ROCK (Tyr40, depending on the type of mutation generated) family members. The Tyr40 residue is also involved in the engagement of the cytoskeletal regulated serum response factor, stress fiber induction, and focus formation in immortalized fibroblasts [148]. These mutants can be referred to as ‘signaling branch deficient’, since they still conserve the ability to interact with other downstream elements [148]. (c) Gene alterations that lead to the elimination of RHOA expression. All these mutations are usually found in heterozygosis in patients, although the concurrent loss of the wild-type allele is also frequently observed in the case of diffuse-type gastric cancer [30]. Other tumor-associated mutations found in genes encoding RHOA signaling elements that are paradoxical according to the accepted functional archetype for these GTPases can be also probably explained according to this new putative RHOA tumor suppression roles. Those include the recurrent loss of genes encoding RHOA GEFs (ARHGEF10 and ARHGEF10L) [149], the gain-of-function mutations found in RHOA GAP-encoding genes (e.g. ARHGAP6, ARHGAP26 and ARHGAP35) [41,144,150], and the loss-of-function mutations detected in genes encoding myosin heavy chains (e.g. MYH9, MYH10 and MYH14) [151,152] (further information can be searched at the inTOgen database using the link: https://www.intogen.org/search) (Figure 1).
Evidence supporting these new antitumorigenic roles has been obtained using several animal models. Thus, the analysis of CRISPR/Cas9-genome edited mice expressing RhoAG17V from the endogenous RhoA locus has clearly established the role of this dominant negative mutant in T-cell lymphomagenesis [153] (Figure 3). The same results were obtained in leukemogenic experiments using mice reconstituted with RhoAG17V-expressing bone marrow precursors [154]. In both cases, however, this function could be only triggered upon the concurrent genetic elimination of Tet2, a transcriptional repressor that is usually mutated at the same time that RHOA in T-cell lymphomas [153,154]. Although the signaling involved is still unknown, available data suggest that RhoAG17V favors this tumorigenic process by activating the PI3K–AKT–mTOR axis in an ICOS receptor-dependent manner [153,154]. This, in turn, favors the differentiation and proliferation of the preneoplasic cells towards the follicular helper T cell lineage [153]. Interestingly, this is the T-cell lineage whose alteration leads to the formation of this type of leukemia in patients [153]. The expression of another dominant negative version, RhoAT19N, has been also shown to favor both Apc+/− (adenomatous polyposis coli)-driven colorectal cancer and metastasis. This protumorigenic effect is mediated in this case by the elevation of the signaling output from the WNT–β-catenin pathway [94] (Figure 3), suggesting that the effects of the RHOA dominant negative mutants are tumor type-specific. Other animal models have also demonstrated the implication of RHO signaling elements in cell transformation. Hence, the use of a Zebrafish model has demonstrated that the expression of the dominant negative (RHOAT19N) and constitutively active (RHOAQ63L) versions of RHOA promotes and abates KRAS oncogene-driven liver tumorigenesis, respectively [155]. As in the case of leukemia, the signaling pathways involved in these biological programs remain to be elucidated. Genetic analysis using genetically modified mice also indicates that the elimination of either RhoA or Myh9 favors tumorigenesis in many tissues [97,152,156–158] (Figures 1 and 3). Conversely, the regulatory subunits of the phosphatases that inactivate the myosin light chain behave as oncogenic drivers when tested in breast cancer models [152] (Figure 1). In the case of skin, it has been proposed that the effect of the loss of RhoA is linked to increased proliferation of keratinocytes due to elevated abundance of the RhoB GTPase [156], although this seems prima facie at odds with the tumor suppressor functions described for RhoB in the same tumorigenic protocols [159]. In the case of the loss of myosin heavy chain IIA (encoded by the Myh9 gene), there are conflicting reports on the possible mechanism involved as defects associated with the loss of p53 and the induction of genomic instability due to frequent alterations in cytokinesis have been postulated [157,158].
It is as yet unclear whether all these loss-of-function mutations in the RHOA pathway elicit the same pathobiological and clinical effects (Figure 4A). Given that it should be much easier to generate any random mutation that leads to the elimination of the protein, the detection of very specific RHOA point mutations in tumors suggests that they must confer some unknown Darwinian selective advantage that cannot be provided by the null mutations. Consistent with this idea, it has been shown that the signaling and pathogenic effects elicited by RhoAG17V in mouse lymphocytes cannot be recapitulated by the genetic deletion of the wild-type Rhoa locus [153]. We can also surmise that the dominant negative RHOA mutants will elicit wider signaling effects in cells than the signaling branch-deficient counterparts due to the ability to block upstream RHO GEFs (Figure 4A, compare model a with c), although this idea has not been experimentally validated as yet in primary tumor cells. Another lingering question in this area is the downstream pathways involved in the engagement of these tumor suppression effects (Figure 4A). Members of the PKN family are obvious candidates, since the activation of these serine/threonine kinases will be abolished in the three classes of RHOA mutations described above in most cases [148] (Figure 4A, model c). Another possibility that does not exclude the previous one is that this biological response is mediated by the activation of actomyosin-dependent pathways (Figure 1), an idea supported by the tumor suppressor roles recently assigned to many myosin heavy chains [152,157,158]. This possibility seems at odds with the known proliferative effects induced by constitutively versions of ROCK2 in transgenic mice [65,66] (Figure 3), but is consistent with observations demonstrating that the addition of Y-27632, a drug that blocks ROCK, PKN and related AGC kinases, mimics the effect of ectopically expressed dominant negative versions of RHOA when added to intestinal organoids [30]. We can also speculate that the effects of these RHOA mutants could be just the consequence of the loss of normal biological processes not directly linked to tumor suppression. For example, the transformation associated with the loss of the mouse Myh9 gene has been recently attributed to cytokinesis defects that lead to increased genomic instability in mice [157]. A recent report suggests a more counterintuitive possibility, at least in the case of the dominant negative RHOA mutations found in lymphomas: a neomorphic, gain-of-function effect that can promote the activation of protumorigenic pathways (Figure 4A, point e). This idea derives from the observation that ectopically expressed RHOAG17V can tightly bind to the VAV1 GEF, a property that favors the RHOAG17V-mediated tethering of this GEF to the plasma membrane, its phosphorylation-mediated activation, and the subsequent engagement of VAV1-dependent signaling pathways [160]. Although the physiological significance of this mechanism remains to be corroborated in patient-derived tumor cells, the foregoing results remind us that we must approach the function of these unexpected RHOA pathway mutants with totally open minds. As in the case of the gain-of-function mutations, current data from genetically manipulated mice indicate that both the dominant negative and null RHOA gene mutations can only drive primary tumorigenesis when combined with additional genetic lesions [94,97,153,154,156]. This is consistent with the development of RHOA mutations in late stages of human tumors [137].
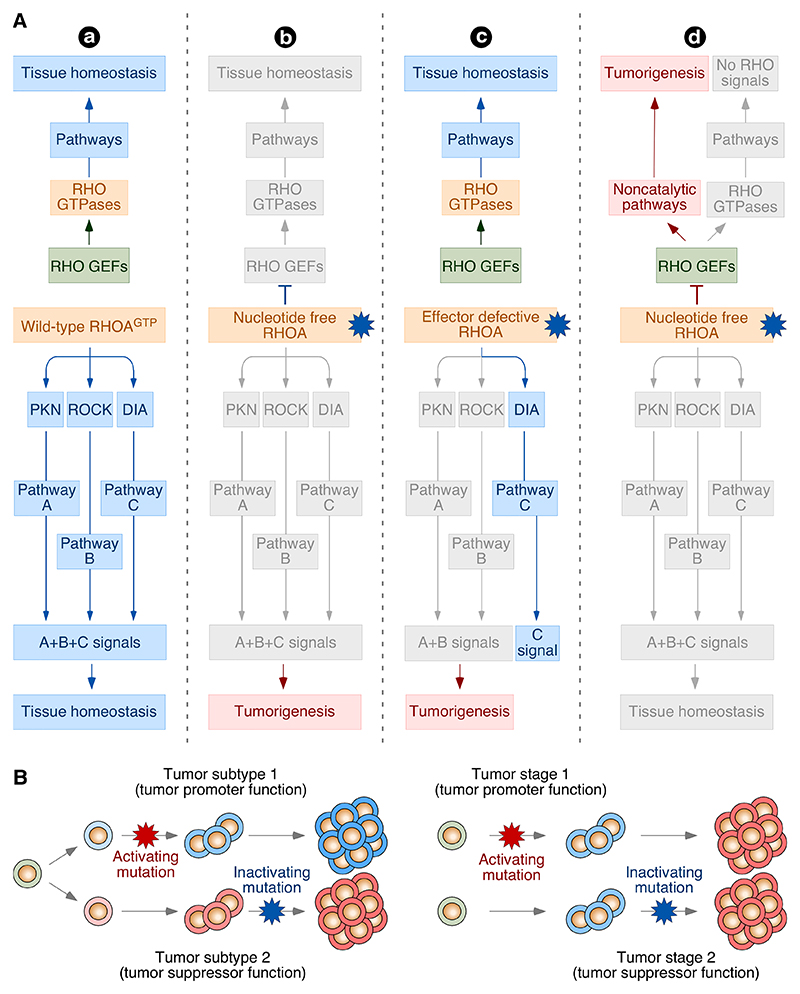
Figure 4. Type of putative alterations in loss- and gain-of-function mutations in RHO signaling elements.
(A) Potential signaling effects induced by wild-type (a), nucleotide-free mutant (b and d), and signaling branch defective mutant (c) versions of RHOA in cells. In model b, the expression of the dominant negative RHOA mutant leads to the disruption of downstream signaling that can favor tumorigenesis by either the elimination of tumor suppressor functions or the dysfunction of normal biological processes. In addition, the binding of this mutant to upstream RHO GEFs can disrupt the signaling of the protein product of the wild-type RHOA allele present in cancer cells and of additional GTPase substrates of those RHOA GEFs. This latter defect should not occur in the case of signaling branch-deficient RHOA mutants (model c). These latter mutants should also have a minor impact on the overall downstream signaling of the normal protein (model c). In model d, the expression of the nucleotide-free RHOA mutant protein elicits a neomorphic, gain-of-function effect on upstream RHO GEFs. This model does not exclude the cooperativity between these pathways and the defective RHOA signaling proposed in the other models. This possibility was not included for the sake of simplicity. (B) Depiction of the possible segregation of the tumor promoter and suppression functions of RHO pathways in either clinically different subtypes of the same tumor (left) or in different progression stages of the same tumor type (right) according to current experimental evidence. See further details in the main text. In A and B, loss- and gain-of-function mutations are depicted as blue and red 10 point stars, respectively.
Before the identification of these RHOA mutations, the geranylgeranylated version of RHOB was the only classical RHO family member known to be associated with tumor suppressor activities [15]. However, recent data suggest that such activities can be also expanded to other RHO subgroups. Thus, PREX2 and PAK3 have been found recurrently deleted in pancreatic cancer and desmoplastic melanoma, respectively [25,161] (Figure 1). Signaling studies also suggest that TIAM1 might participate in the suppression of colorectal cancer by inhibiting the protumorigenic YAP/TAZ pathway [115]. At this moment, however, the relevance of these pathways in vivo remains to be validated using animal models. Finally, a study has reported that CDC42 protects against the spontaneous development of hepatocellular carcinoma in mice [162], an observation that further suggests a potential tumor suppressor function for this GTPase as well. However, mutations in this gene are very rare in the tumors that have been sequenced so far (Figure 2A). Taking together, these observations suggest that we are only seeing the tip of the iceberg regarding the implication of RHO-dependent pathways in tumor suppression.
GTPase-independent functions of RHO signaling elements in tumorigenesis
Adding further complexity to the contribution of RHO proteins to the biology of cancer cells, recent observations indicate that RHO signaling elements can also trigger GTPase-independent pro- and antitumorigenic functions. Examples of the former ones include the catalysis-independent stimulation of: (a) the K-RAS–RAF–ERK pathway by ARHGEF2 (also known as GEF-H1) [163]. (b) The K-RAS–PI3K–AKT axis by P-REX2 [164]. (c) The YAP/TAZ complex by ARHGEF7 (also referred to as β-PIX) [165]. (d) The nuclear factor of activated T cells by VAV1 [166]. (e) The estrogen receptor by VAV3 [167]. This pathway is even less canonical than the others included in this list for it requires the nuclear localization of this GEF [168]. RAC1 itself can be also included here as it can stimulate mTOR signaling using a protein–protein interaction-based mechanism that takes place independently of the switch regions and the type of guanosine nucleotide state of the GTPase [169]. Examples for potential catalysis-independent tumor suppression functions include the ARHGEF3 (also known as XPLN and RHO GEF3)- and the VAV1-mediated inhibition of mTORC2 and the active cytosolic fragment of the NOTCH1 oncoprotein, respectively [170,171]. The tumor suppression roles in the latter pathway in T-cell acute lymphoblastic leukemia (T-ALL) have been recently demonstrated using both mouse models and patient-derived T-ALL cells [171]. They also probably operate in other tumors, as inferred by the presence of VAV1 mutations that generate proteins that cannot promote the degradation of NOTCH1 [172]. This probably explains the detection of both gain- and loss-of-function mutations in this gene in a variety of tumors [172] (Figure 1). Additional noncatalytic, adaptor-like functions have been described for RHOA GAPs (DLC family members, ARHGAP36) [56,173] and downstream effectors (PAK1) [174,175]. These noncanonical functions are probably more widespread, given that the catalytic domains of some RHO GEFs (e.g. ARHGEF7) and GAPs (e.g. ARHGAP36, SRGAP3) show no detectable enzyme activity. Some of these signaling elements also have isoforms generated by differential splicing that lack the catalytic domains (e.g. the short isoform of the GEF VAV3).
Organization of tumor promoting and suppressing functions
The roles of RHO signaling elements in tumor promotion and suppression raise the issue of how these antagonistic functions are segregated in tumors. In addition to its obvious academic interest, the elucidation of this issue is critical to establish the therapeutic viability of RHO pathways. Although little information exists as yet, we can infer some organizational possibilities from current experimental evidence. These two functions seem to be separated in some cases in different subtypes of the same tumor (Figure 4B, model on the left). For example, the loss- and gain-of-function RHOA mutations are preferentially found in lymphoma patients belonging to the memory and regulatory T-cell subtypes, respectively [53]. Similarly, the antitumorigenic and tumorigenic roles of VAV1 in T-cell tumors are segregated between immature and mature T cells, respectively [171] (unpublished observations from our laboratory). On the other hand, data from carcinogen-induced tumorigenesis experiments using Tiam1−/− and Rhob−/− knockout mice suggest that the pro- and antitumorigenic roles of these proteins could be segregated in early and late phases of skin tumors, respectively (Figure 4B, model on the right). Accordingly, these two mouse strains display lower disease burden due to defects in tumor initiation but, on the other hand, the few tumors that eventually develop show more aggressive features than those from wild-type counterparts [99]. Bivalent functions of the RAC GAP β2-chimerin have also been recently described along the progression of breast tumors [82]. Tiam1 exhibits functions that could explain this late-phase tumorigenic effects, including those related to chromosomal stability and epithelial integrity [112,113,176,177]. It is worth noting, however, that these data can be alternatively explained by Darwinian selection events that could favor the emergence of alternative pathways to compensate for the absence of Tiam1 and RhoB during the tumor initiation phase. It will be interesting to generate in the near-future inducible knock-in mice to better dissect the orchestration of these disparate functions of RHO pathways in tumorigenic processes.
RHO pathway-based therapeutics
Given their roles in tumorigenesis, there has been a historical interest in the development of drug inhibitors for RHO signaling elements. Drug screenings carried out during the last decade have led, in fact, to a large menagerie of chemical probes that target the maturation of RHO GTPases, the interaction of RHO GTPases with upstream RHO GEFs, the catalytic activity of RHO GEFs, and the enzyme activity of downstream signaling elements [178] (Figure 5). Although these data look promising, we should be aware that they are associated with many drawbacks. One of them is that many of those inhibitors have been developed without taken into consideration the actual viability of them outside academic settings. Thus, inhibitors for geranylgeranyl transferases have been extensively pursued despite the known fact that these enzymes contribute to the posttranslational modification of a large plethora of intracellular substrates. Likewise, inhibitory compounds for RHO GTPases have been isolated despite available data from knockout mice, indicating that such inhibition would be probably linked to unavoidable toxic effects [1]. Intense efforts have also been focused on RHO GEF inhibitors despite the widely accepted poor druggability of their catalytic domains [11]. Very limited efforts have been also devoted to determining the toxicity and target specificity of most of those compounds in vivo. Another draw-back is the chronic lack of adequate mouse models in this field that could allow us to establish the usefulness and collateral side effects of the systemic inhibition of the selected drug targets in fully formed tumors. Last but not least, the recently described tumor suppression activities of RHO signaling elements raise concerns regarding the general application of RHO pathway inhibitors without the proper identification of the specific roles they play in each tumor type.
Figure 5. Pharmacological inhibition of RHO-regulated pathways.
Inhibitors targeting the indicated regulatory and effector steps are shown in light red speech bubbles. Secramine only works on CDC42, favoring its interaction with RHO GDIs. EHT1864 traps RHO proteins in the GDP-bound form. The rest of compounds are inhibitory. The list of PI3Kβ drugs includes both isoform-specific and pan-specific compounds.
Notwithstanding these Damocles’ swords, it is obvious that the targeting of RHO pathways is still a potentially interesting therapeutic strategy for cancer and other epidemiologically relevant diseases. In my view, the most feasible drug targets are the RHO signaling elements that show intrinsic catalytic activity. In this category, an obvious option is still the upstream RHO GEFs (Figure 5). The catalytic domain of these enzymes is not easily druggable as discussed above. However, some GEFs display both multidomain catalytic cassettes and allosteric mechanisms of activation (e.g. VAV family proteins and ARHGEF12) that can be used for the development of high-affinity and specific inhibitors targeting pockets outside the GEF–GTPase-binding interface. The recent isolation of inhibitors (Y16 and derivatives) that dock onto a groove present between the catalytic and adjacent pleckstrin homology domain of ARHGEF12 demonstrates that this type of avenues is possible for some GEFs [179] (Figure 5). Tool kits to inhibit these enzymes can be wider than chemical drugs in the near future, as evidenced by the recent development of both peptide (TRIPE32G) and RNA (K11, K91) aptamers that can inhibit TRIO and TIAM1, respectively [180,181] (Figure 5). Effective efforts in any of those directions, however, will need a better understanding of the 3D structure of those GEFs. These therapies are potentially interesting given that, unlike the case of RHO GTPases, the systemic inhibition of most RHO GEFs does not usually lead to dire toxic effects. However, in view of recent data, we must clarify in advance the implication of each of the potential target GEFs in the regulation of catalysis-dependent tumor suppression mechanisms. Other obvious options are RHO downstream kinases such as PAK and ROCK family proteins (Figure 5), although the analyses of mouse models suggest that some of them might be associated with unavoidable toxicity [47]. An extensive collection of ATP competitive and noncompetitive inhibitors for PAK family members has been already developed although, so far, none of them have reached the clinic due to chemical instability, poor biocompatibility or high toxicity [47,178] (Figure 5). A large number of ATP competitive inhibitors for ROCK family members have also been developed (Figure 5). These inhibitors do not seem to be toxic, although they are, in general, highly unspecific as they can target many other related kinases. Some of these inhibitors have moved into the clinic to treat cerebral vasospasms, pulmonary hypertension and glaucoma, although none of them have been approved as cancer therapies [54,178]. Further efforts in this area will probably result in more specific and biocompatible drugs for these and other RHO downstream effectors in the near future. However, as in the case of the GEFs, we must clarify before whether these inhibitors could lead to the unwanted inactivation of tumor suppression mechanism as inferred from the results with the Y-27632 inhibitor in some organoid models [30]. Another complementary, long-term avenue is to dig deeper into the most distal downstream elements of the RHO-dependent pathways to identify new druggable targets. For example, the analysis of the VAV family-dependent transcriptome has led to the identification of druggable downstream targets whose inactivation prompted defects in the fitness of breast cancer cells similar to those found upon the depletion of VAV family proteins [81]. Additional targets could be discovered when the still limited understanding of the contribution of RHO pathways to the transcriptome of cancer cells is expanded in the near future. Considering the recently discovered variegated roles of RHO pathways in cancer, the implementation of any those potential therapies will necessarily involve a prior stratification of patients to limit their application to those where the RHO pathways play pro-rather than antitumorigenic roles. It is also likely that those drugs will have to be used in combinatorial therapies, given the intrinsic mutational complexity of most tumors. Evidence for the synergistic effects of the concurrent inhibition of RHO pathway with K-RAS signaling elements, cell cycle regulators, and standard chemotherapy agents is already available in preclinical models [118,120,129,131,132,182].
Concluding remarks
Significant advances have been made during these last years in the understanding of the modus operandi of RHO GTPases in cancer. Despite this, we still have a long way to go to clarify many pending questions such as the functional and clinical significance of many of the mutations found in tumors, the pathways that contribute to cancer development, the organization of the RHO tumor promoter and suppression functions in different cancers, and the identification of the best Achilles’ heels to target pharmacologically within those pathways. To this end, we will need to implement better animal models and more close-to-the-clinic experimental tools such as patient-derived xenografts, organoids, and primary cancer cells. Finally, the development of better pharma-comimetic mouse models, the selection of wider ranges of druggable targets, a better understanding of the 3D structures of the selected targets, the development of alternative pharmacological tool kits, and further screening methods should also allow us to reach the holy grail of the development of effective therapies in the near future.
Acknowledgements
The author likes to thank M. Dosil for her comments on the manuscript.
Funding
My work is supported by grants from the Castilla-León Government [CSI049U16], Spanish Ministry of Economy and Competitiveness [SAF2015-64556-R], Worldwide Cancer Research [14-1248], Ramón Areces Foundation, and Spanish Society against Cancer [GC16173472GARC]. Funding from Spanish national and regional governments is partially contributed by the European Regional Development Fund.
Abbreviations
- APC
adenomatous polyposis coli
- Dock
dedicator of cytokinesis
- GAPs
GTPase-activating proteins
- GDIs
GDP dissociation inhibitors
- GDS
GDP dissociation stimulator
- GEFs
GDP/GTP exchange factors
- PAK
p21-activated kinase
- PI3K
phosphatidylinositol 3-phosphate kinase
- PKN
protein kinase N
- ROCK
RHO-associated coiled-coil containing protein kinase
- T-ALL
T-cell acute lymphoblastic leukemia.
Footnotes
Competing Interests
The Author declares that there are no competing interests associated with this manuscript.
References
- 1.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. BioEssays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 3.Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 4.Loirand G, Pacaud P. The role of Rho protein signaling in hypertension. Nat Rev Cardiol. 2010;7:637–647. doi: 10.1038/nrcardio.2010.136. [DOI] [PubMed] [Google Scholar]
- 5.Fryer BH, Field J. Rho, Rac, Pak and angiogenesis: old roles and newly identified responsibilities in endothelial cells. Cancer Lett. 2005;229:13–23. doi: 10.1016/j.canlet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol. 2004;5:355–366. doi: 10.1038/nrm1365. [DOI] [PubMed] [Google Scholar]
- 7.Bustelo XR. Understanding Rho/Rac biology in T-cells using animal models. BioEssays. 2002;24:602–612. doi: 10.1002/bies.10107. [DOI] [PubMed] [Google Scholar]
- 8.Mulloy JC, Cancelas JA, Filippi MD, Kalfa TA, Guo F, Zheng Y. Rho GTPases in hematopoiesis and hemopathies. Blood. 2010;115:936–947. doi: 10.1182/blood-2009-09-198127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newey SE, Velamoor V, Govek EE, Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64:58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- 10.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 11.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter AP, Papaioannou A, Malliri A. Deregulation of Rho GTPases in cancer. Small GTPases. 2016;7:123–138. doi: 10.1080/21541248.2016.1173767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zandvakili I, Lin Y, Morris JC, Zheng Y. Rho GTPases: anti-or pro-neoplastic targets? Oncogene. 2017;36:3213–3222. doi: 10.1038/onc.2016.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 15.Du W, Lebowitz PF, Prendergast GC. Cell growth inhibition by farnesyltransferase inhibitors is mediated by gain of geranylgeranylated RhoB. Mol Cell Biol. 1999;19:1831–1840. doi: 10.1128/MCB.19.3.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams CL. A new signaling paradigm to control the prenylation and trafficking of small GTPases. Cell Cycle. 2013;12:2933–2934. doi: 10.4161/cc.26230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bustelo XR, Ojeda V, Barreira M, Sauzeau V, Castro-Castro A. Rac-ing to the plasma membrane: the long and complex work commute of Rac1 during cell signaling. Small GTPases. 2012;3:60–66. doi: 10.4161/sgtp.19111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lerida I, Muriel O, et al. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 2011;30:3913–3927. doi: 10.1038/emboj.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castro-Castro A, Muriel O, Del Pozo MA, Bustelo XR. Characterization of novel molecular mechanisms favoring Rac1 membrane translocation. PLoS ONE. 2016;11:e0166715. doi: 10.1371/journal.pone.0166715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 21.Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, et al. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153:1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, Bi F, Zhou X, Zheng Y. Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol. 2012;22:365–373. doi: 10.1016/j.tcb.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawazu M, Ueno T, Kontani K, Ogita Y, Ando M, Fukumura K, et al. Transforming mutations of RAC guanosine triphosphatases in human cancers. Proc Natl Acad Sci USA. 2013;110:3029–3034. doi: 10.1073/pnas.1216141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet. 2015;47:1194–1199. doi: 10.1038/ng.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 27.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33:125–136.:e3. doi: 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 31.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luc GT, Morris LG, Chandramohan R, West L, Zehir A, Chakravarty D, Pfister DG, et al. The molecular landscape of recurrent and metastatic head and neck cancers: insights from a precision oncology sequencing platform. JAMA Oncol. 2017;3:244–255. doi: 10.1001/jamaoncol.2016.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.:e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540–556.:e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Chang MT, Johnsen HC, Gao SP, Sylvester BE, Sumer SO, et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med. 2017;9:4. doi: 10.1186/s13073-016-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abate F, da Silva-Almeida AC, Zairis S, Robles-Valero J, Couronne L, Khiabanian H, et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc Natl Acad Sci USA. 2017;114:764–769. doi: 10.1073/pnas.1608839114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 42.Boddicker RL, Razidlo GL, Dasari S, Zeng Y, Hu G, Knudson RA, et al. Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma. Blood. 2016;128:1234–1245. doi: 10.1182/blood-2016-03-707141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallois D, Dobay MP, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128:1490–1502. doi: 10.1182/blood-2016-02-698977. [DOI] [PubMed] [Google Scholar]
- 44.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47:1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 45.Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:371–375. doi: 10.1038/ng.2916. [DOI] [PubMed] [Google Scholar]
- 46.Srijakotre N, Man J, Ooms LM, Lucato CM, Ellisdon AM, Mitchell CA. P-Rex1 and P-Rex2 RacGEFs and cancer. Biochem Soc Trans. 2017;45:963–977. doi: 10.1042/BST20160269. [DOI] [PubMed] [Google Scholar]
- 47.Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014;14:13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen EMV, Mouw KW, Kim P, Iyer G, Wagle N, Al-Ahmadie H, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lochhead PA, Wickman G, Mezna M, Olson MF. Activating ROCK1 somatic mutations in human cancer. Oncogene. 2010;29:2591–2598. doi: 10.1038/onc.2010.3. [DOI] [PubMed] [Google Scholar]
- 52.Park H, Cho SY, Kim H, Na D, Han JY, Chae J, et al. Genomic alterations in BCL2L1 and DLC1 contribute to drug sensitivity in gastric cancer. Proc Natl Acad Sci USA. 2015;112:12492–12497. doi: 10.1073/pnas.1507491112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagata Y, Kontani K, Enami T, Kataoka K, Ishii R, Totoki Y, et al. Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood. 2016;127:596–604. doi: 10.1182/blood-2015-06-644948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13:900–908. doi: 10.1038/embor.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tala I, Chen R, Hu T, Fitzpatrick ER, Williams DA, Whitehead IP. Contributions of the RhoGEF activity of p210 BCR/ABL to disease progression. Leukemia. 2013;27:1080–1089. doi: 10.1038/leu.2012.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barras D, Widmann C. GAP-independent functions of DLC1 in metastasis. Cancer Met Rev. 2014;33:87–100. doi: 10.1007/s10555-013-9458-0. [DOI] [PubMed] [Google Scholar]
- 57.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large b cell lymphoma. Cell. 2017;171:481–494.:e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc Natl Acad Sci USA. 2013;110:912–917. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7:794–797. doi: 10.1016/S0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- 62.Shakir MA, Gill JS, Lundquist EA. Interactions of UNC-34 enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics. 2006;172:893–913. doi: 10.1534/genetics.105.046359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dalton LE, Kamarashev J, Barinaga-Rementeria Ramirez I, White G, Malliri A, Hurlstone A. Constitutive RAC activation is not sufficient to initiate melanocyte neoplasia but accelerates malignant progression. J Invest Dermatol. 2013;133:1572–1581. doi: 10.1038/jid.2013.23. [DOI] [PubMed] [Google Scholar]
- 64.Lissanu Deribe Y, Shi Y, Rai K, Nezi L, Amin SB, Wu CC, et al. Truncating PREX2 mutations activate its GEF activity and alter gene expression regulation in NRAS-mutant melanoma. Proc Natl Acad Sci USA. 2016;113:E1296–E1305. doi: 10.1073/pnas.1513801113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, et al. Actomyosin-mediated cellular tension drives increased tissue stiffness and beta-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell. 2011;19:776–791. doi: 10.1016/j.ccr.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masre SF, Rath N, Olson MF, Greenhalgh DA. ROCK2/ras(Ha) co-operation induces malignant conversion via p53 loss, elevated NF-κB and tenascin C-associated rigidity, but p21 inhibits ROCK2/NF-κB-mediated progression. Oncogene. 2017;36:2529–2542. doi: 10.1038/onc.2016.402. [DOI] [PubMed] [Google Scholar]
- 67.van der Horst EH, Degenhardt YY, Strelow A, Slavin A, Chinn L, Orf J, et al. Metastatic properties and genomic amplification of the tyrosine kinase gene ACK1. Proc. Natl Acad Sci USA. 2005;102:15901–15906. doi: 10.1073/pnas.0508014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Climente-Gonzalez H, Porta-Pardo E, Godzik A, Eyras E. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20:2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 69.Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene. 1999;18:6835–6839. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 70.Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, et al. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene. 2000;19:3013–3020. doi: 10.1038/sj.onc.1203621. [DOI] [PubMed] [Google Scholar]
- 71.Singh A, Karnoub AE, Palmby TR, Lengyel E, Sondek J, Der CJ. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene. 2004;23:9369–9380. doi: 10.1038/sj.onc.1208182. [DOI] [PubMed] [Google Scholar]
- 72.Zhou C, Licciulli S, Avila JL, Cho M, Troutman S, Jiang P, et al. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene. 2013;32:903–909. doi: 10.1038/onc.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stallings-Mann ML, Waldmann J, Zhang Y, Miller E, Gauthier ML, Visscher DW, et al. Matrix metalloproteinase induction of Rac1b, a key effector of lung cancer progression. Sci Transl Med. 2012;4:142ra95. doi: 10.1126/scitranslmed.3004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He X, Yuan C, Yang J. Regulation and functional significance of CDC42 alternative splicing in ovarian cancer. Oncotarget. 2015;6:29651–29663. doi: 10.18632/oncotarget.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshizuka N, Moriuchi R, Mori T, Yamada K, Hasegawa S, Maeda T, et al. An alternative transcript derived from the trio locus encodes a guanosine nucleotide exchange factor with mouse cell-transforming potential. J Biol Chem. 2004;279:43998–44004. doi: 10.1074/jbc.M406082200. [DOI] [PubMed] [Google Scholar]
- 76.Trenkle T, McClelland M, Adlkofer K, Welsh J. Major transcript variants of VAV3, a new member of the VAV family of guanine nucleotide exchange factors. Gene. 2000;245:139–149. doi: 10.1016/S0378-1119(00)00026-3. [DOI] [PubMed] [Google Scholar]
- 77.Reimer D, Boesch M, Wolf D, Marth C, Sopper S, Hatina J, et al. Truncated isoform Vav3.1 is highly expressed in ovarian cancer stem cells and clinically relevant in predicting prognosis and platinum-response. Int J Cancer. 2018;142:1640–1651. doi: 10.1002/ijc.31186. [DOI] [PubMed] [Google Scholar]
- 78.Trenkle T, Hakim SG, Jacobsen HC, Sieg P. Differential gene expression of the proto-oncogene VAV3 and the transcript variant VAV3.1 in oral squamous cell carcinoma. Anticancer Res. 2015;35:2593–2600. [PubMed] [Google Scholar]
- 79.Hakem A, Sanchez-Sweatman O, You-Ten A, Duncan G, Wakeham A, Khokha R, et al. Rhoc is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strumane K, Rygiel T, van der Valk M, Collard JG. Tiam1-deficiency impairs mammary tumor formation in MMTV-c-neu but not in MMTV-c-myc mice. J Cancer Res Clin Oncol. 2009;135:69–80. doi: 10.1007/s00432-008-0437-8. [DOI] [PubMed] [Google Scholar]
- 81.Citterio C, Menacho-Marquez M, Garcia-Escudero R, Larive RM, Barreiro O, Sanchez-Madrid F, et al. The Rho exchange factors Vav2 and Vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Sci Signal. 2012;5:ra71. doi: 10.1126/scisignal.2002962. [DOI] [PubMed] [Google Scholar]
- 82.Casado-Medrano V, Barrio-Real L, Garcia-Rostan G, Baumann M, Rocks O, Caloca MJ. A new role of the Rac-GAP beta2-chimaerin in cell adhesion reveals opposite functions in breast cancer initiation and tumor progression. Oncotarget. 2016;7:28301–28319. doi: 10.18632/oncotarget.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Basak P, Leslie H, Dillon RL, Muller WJ, Raouf A, Mowat MRA. In vivo evidence supporting a metastasis suppressor role for Stard13 (Dlc2) in ErbB2 (Neu) oncogene induced mouse mammary tumors. Genes Chromosomes Cancer. 2018;57:182–191. doi: 10.1002/gcc.22519. [DOI] [PubMed] [Google Scholar]
- 84.McHenry PR, Sears JC, Herrick MP, Chang P, Heckman-Stoddard BM, Rybarczyk M, et al. P190b RhoGAP has pro-tumorigenic functions during MMTV-Neu mammary tumorigenesis and metastasis. Breast Cancer Res. 2010;12:R73. doi: 10.1186/bcr2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heckman-Stoddard BM, Vargo-Gogola T, McHenry PR, Jiang V, Herrick MP, Hilsenbeck SG, et al. Haploinsufficiency for p190B RhoGAP inhibits MMTV-Neu tumor progression. Breast Cancer Res. 2009;11:R61. doi: 10.1186/bcr2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arias-Romero LE, Villamar-Cruz O, Huang M, Hoeflich KP, Chernoff J. Pak1 kinase links ErbB2 to beta-catenin in transformation of breast epithelial cells. Cancer Res. 2013;73:3671–3682. doi: 10.1158/0008-5472.CAN-12-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang RA, Zhang H, Balasenthil S, Medina D, Kumar R. PAK1 hyperactivation is sufficient for mammary gland tumor formation. Oncogene. 2006;25:2931–2936. doi: 10.1038/sj.onc.1209309. [DOI] [PubMed] [Google Scholar]
- 88.Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, Bender Kim CF, Sweet-Cordero A, et al. Requirement for Rac1 in a K-Ras induced lung cancer in the mouse. Cancer Res. 2007;67:8089–8094. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 89.Wang Z, Pedersen E, Basse A, Lefever T, Peyrollier K, Kapoor S, et al. Rac1 is crucial for Ras-dependent skin tumor formation by controlling Pak1-Mek-Erk hyperactivation and hyperproliferation in vivo. Oncogene. 2010;29:3362–3373. doi: 10.1038/onc.2010.95. [DOI] [PubMed] [Google Scholar]
- 90.Samuel MS, Lourenco FC, Olson MF. K-Ras mediated murine epidermal tumorigenesis is dependent upon and associated with elevated Rac1 activity. PLoS ONE. 2011;6:e17143. doi: 10.1371/journal.pone.0017143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mizukawa B, Wei J, Shrestha M, Wunderlich M, Chou FS, Griesinger A, et al. Inhibition of Rac GTPase signaling and downstream prosurvival Bcl-2 proteins as combination targeted therapy in MLL-AF9 leukemia. Blood. 2011;118:5235–5245. doi: 10.1182/blood-2011-04-351817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu CY, Carpenter ES, Takeuchi KK, Halbrook CJ, Peverley LV, Bien H, et al. PI3K regulation of RAC1 is required for KRAS-induced pancreatic tumorigenesis in mice. Gastroenterology. 2014;147:1405–1416.:e7. doi: 10.1053/j.gastro.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sakamori R, Yu S, Zhang X, Hoffman A, Sun J, Das S, et al. CDC42 inhibition suppresses progression of incipient intestinal tumors. Cancer Res. 2014;74:5480–5492. doi: 10.1158/0008-5472.CAN-14-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rodrigues P, Macaya I, Bazzocco S, Mazzolini R, Andretta E, Dopeso H, et al. RHOA inactivation enhances Wnt signalling and promotes colorectal cancer. Nat Commun. 2014;5:5458. doi: 10.1038/ncomms6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meyer N, Peyret-Lacombe A, Canguilhem B, Medale-Giamarchi C, Mamouni K, Cristini A, et al. Rhob promotes cancer initiation by protecting keratinocytes from UVB-induced apoptosis but limits tumor aggressiveness. J Invest Dermatol. 2014;134:203–212. doi: 10.1038/jid.2013.278. [DOI] [PubMed] [Google Scholar]
- 96.Frances D, Sharma N, Pofahl R, Maneck M, Behrendt K, Reuter K, et al. A role for Rac1 activity in malignant progression of sebaceous skin tumors. Oncogene. 2015;34:5505–5512. doi: 10.1038/onc.2014.471. [DOI] [PubMed] [Google Scholar]
- 97.Zandvakili I, Davis AK, Hu G, Zheng Y. Loss of RhoA exacerbates, rather than dampens, oncogenic K-Ras induced lung adenoma formation in mice. PLoS ONE. 2015;10:e0127923. doi: 10.1371/journal.pone.0127923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Malliri A, Rygiel TP, van der Kammen RA, Song JY, Engers R, Hurlstone AF, et al. The Rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J Biol Chem. 2006;281:543–548. doi: 10.1074/jbc.M507582200. [DOI] [PubMed] [Google Scholar]
- 99.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 100.Menacho-Marquez M, Garcia-Escudero R, Ojeda V, Abad A, Delgado P, Costa C, et al. The Rho exchange factors Vav2 and Vav3 favor skin tumor initiation and promotion by engaging extracellular signaling loops. PLoS Biol. 2013;11:e1001615. doi: 10.1371/journal.pbio.1001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lindsay CR, Lawn S, Campbell AD, Faller WJ, Rambow F, Mort RL, et al. P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nat Commun. 2011;2:555. doi: 10.1038/ncomms1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Justilien V, Ali SA, Jamieson L, Yin N, Cox AD, Der CJ, et al. Ect2-dependent rRNA synthesis is required for KRAS-TRP53-driven lung adenocarcinoma. Cancer Cell. 2017;31:256–269. doi: 10.1016/j.ccell.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu LN, Zhang QL, Li X, Hua X, Cui YM, Zhang NJ, et al. Tiam1 transgenic mice display increased tumor invasive and metastatic potential of colorectal cancer after 1,2-dimethylhydrazine treatment. PLoS ONE. 2013;8:e73077. doi: 10.1371/journal.pone.0073077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kawasaki Y, Tsuji S, Muroya K, Furukawa S, Shibata Y, Okuno M, et al. The adenomatous polyposis coli-associated exchange factors Asef and Asef2 are required for adenoma formation in Apc(Min/+)mice. EMBO Rep. 2009;10:1355–1362. doi: 10.1038/embor.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang KH, Sanchez-Aguilera A, Shen S, Sengupta A, Madhu MN, Ficker AM, et al. Vav3 collaborates with p190-BCR-ABL in lymphoid progenitor leukemogenesis, proliferation, and survival. Blood. 2012;120:800–811. doi: 10.1182/blood-2011-06-361709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martin H, Mali RS, Ma P, Chatterjee A, Ramdas B, Sims E, et al. Pak and Rac GTPases promote oncogenic KIT-induced neoplasms. J Clin Invest. 2013;123:4449–4463. doi: 10.1172/JCI67509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sabbir MG, Prieditis H, Ravinsky E, Mowat MR. The role of Dlc1 isoform 2 in K-Ras2(G12D) induced thymic cancer. PLoS ONE. 2012;7:e40302. doi: 10.1371/journal.pone.0040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McDonald ER, III, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. 2017;170:577–592.:e10. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 109.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170:564–576.:e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gonyo P, Bergom C, Brandt AC, Tsaih SW, Sun Y, Bigley TM, et al. SmgGDS is a transient nucleolar protein that protects cells from nucleolar stress and promotes the cell cycle by regulating DREAM complex gene expression. Oncogene. 2017;36:6873–6883. doi: 10.1038/onc.2017.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Whalley HJ, Porter AP, Diamantopoulou Z, White GR, Castaneda-Saucedo E, Malliri A. Cdk1 phosphorylates the Rac activator Tiam1 to activate centrosomal Pak and promote mitotic spindle formation. Nat Commun. 2015;6:7437. doi: 10.1038/ncomms8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woodcock SA, Rushton HJ, Castaneda-Saucedo E, Myant K, White GR, Blyth K, et al. Tiam1-Rac signaling counteracts Eg5 during bipolar spindle assembly to facilitate chromosome congression. Curr Biol. 2010;20:669–675. doi: 10.1016/j.cub.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.David MD, Petit D, Bertoglio J. The RhoGAP ARHGAP19 controls cytokinesis and chromosome segregation in T lymphocytes. J Cell Sci. 2014;127(Pt 2):400–410. doi: 10.1242/jcs.135079. [DOI] [PubMed] [Google Scholar]
- 115.Diamantopoulou Z, White G, Fadlullah MZH, Dreger M, Pickering K, Maltas J, et al. TIAM1 antagonizes TAZ/YAP both in the destruction complex in the cytoplasm and in the nucleus to inhibit invasion of intestinal epithelial cells. Cancer Cell. 2017;31:621–634.:e6. doi: 10.1016/j.ccell.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vu HL, Rosenbaum S, Purwin TJ, Davies MA, Aplin AE. RAC1 p29s regulates PD-L1 expression in melanoma. Pigment Cell Melanoma Res. 2015;28:590–598. doi: 10.1111/pcmr.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bar-Sagi D, Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–238. doi: 10.1016/S0092-8674(00)00115-X. [DOI] [PubMed] [Google Scholar]
- 118.Lu H, Liu S, Zhang G, Bin W, Zhu Y, Frederick DT, et al. PAK signalling drives acquired drug resistance to MAPK inhibitors in BRAF-mutant melanomas. Nature. 2017;550:133–136. doi: 10.1038/nature24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Watson IR, Li L, Cabeceiras PK, Mahdavi M, Gutschner T, Genovese G, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Walsh K, McKinney MS, Love C, Liu Q, Fan A, Patel A, et al. PAK1 mediates resistance to PI3K inhibition in lymphomas. Clin Cancer Res. 2013;19:1106–1115. doi: 10.1158/1078-0432.CCR-12-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Akunuru S, Palumbo J, Zhai QJ, Zheng Y. Rac1 targeting suppresses human non-small cell lung adenocarcinoma cancer stem cell activity. PLoS ONE. 2011;6:e16951. doi: 10.1371/journal.pone.0016951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rao J, Zhou ZH, Yang J, Shi Y, Xu SL, Wang B, et al. Semaphorin-3F suppresses the stemness of colorectal cancer cells by inactivating Rac1. Cancer Lett. 2015;358:76–84. doi: 10.1016/j.canlet.2014.12.040. [DOI] [PubMed] [Google Scholar]
- 123.Lai YJ, Tsai JC, Tseng YT, Wu MS, Liu WS, Lam HI, et al. Small G protein Rac GTPases regulate the maintenance of glioblastoma stem-like cells in vitro and in vivo. Oncotarget. 2017;8:18031–18049. doi: 10.18632/oncotarget.14949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. CellStemCell. 2013;12:761–773. doi: 10.1016/j.stem.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sengupta A, Arnett J, Dunn S, Williams DA, Cancelas JA. Rac2 GTPase deficiency depletes BCR-ABL+ leukemic stem cells and progenitors in vivo. Blood. 2010;116:81–84. doi: 10.1182/blood-2009-10-247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Capala ME, Maat H, Bonardi F, van den Boom V, Kuipers J, Vellenga E, et al. Mitochondrial dysfunction in human leukemic stem/progenitor cells upon loss of RAC2. PLoS ONE. 2015;10:e0128585. doi: 10.1371/journal.pone.0128585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iglesias-Bartolome R, Gutkind JS. Signaling circuitries controlling stem cell fate: to be or not to be. Curr Opin Cell Biol. 2011;23:716–723. doi: 10.1016/j.ceb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Man J, Shoemake J, Zhou W, Fang X, Wu Q, Rizzo A, et al. Sema3c promotes the survival and tumorigenicity of glioma stem cells through Rac1 activation. Cell Rep. 2014;9:1812–1826. doi: 10.1016/j.celrep.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aboukameel A, Muqbil I, Senapedis W, Baloglu E, Landesman Y, Shacham S, et al. Novel p21-activated kinase 4 (PAK4) allosteric modulators overcome drug resistance and stemness in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2017;16:76–87. doi: 10.1158/1535-7163.MCT-16-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aguilar H, Urruticoechea A, Halonen P, Kiyotani K, Mushiroda T, Barril X, et al. VAV3 mediates resistance to breast cancer endocrine therapy. Breast Cancer Res. 2014;16:R53. doi: 10.1186/bcr3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hofbauer SW, Krenn PW, Ganghammer S, Asslaber D, Pichler U, Oberascher K, et al. Tiam1/Rac1 signals contribute to the proliferation and chemoresistance, but not motility, of chronic lymphocytic leukemia cells. Blood. 2014;123:2181–2188. doi: 10.1182/blood-2013-08-523563. [DOI] [PubMed] [Google Scholar]
- 132.Wen X, Huang A, Liu Z, Liu Y, Hu J, Liu J, et al. Downregulation of ROCK2 through nanocomplex sensitizes the cytotoxic effect of temozolomide in U251 glioma cells. PLoS ONE. 2014;9:e92050. doi: 10.1371/journal.pone.0092050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rosenthal DT, Zhang J, Bao L, Zhu L, Wu Z, Toy K, et al. Rhoc impacts the metastatic potential and abundance of breast cancer stem cells. PLoS ONE. 2012;7:e40979. doi: 10.1371/journal.pone.0040979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Islam M, Sharma S, Teknos TN. Rhoc regulates cancer stem cells in head and neck squamous cell carcinoma by overexpressing IL-6 and phosphorylation of STAT3. PLoS ONE. 2014;9:e88527. doi: 10.1371/journal.pone.0088527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 136.Kumar RD, Swamidass SJ, Bose R. Unsupervised detection of cancer driver mutations with parsimony-guided learning. Nat Genet. 2016;48:1288–1294. doi: 10.1038/ng.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cho SY, Park JW, Liu Y, Park YS, Kim JH, Yang H, et al. Sporadic early-onset diffuse gastric cancers have high frequency of somatic cdh1 alterations, but low frequency of somatic RHOA mutations compared with late-onset cancers. Gastroenterology. 2017;153:536–549.:e26. doi: 10.1053/j.gastro.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Janjigian YY, Sanchez-Vega F, Jonsson P, Chatila WK, Hechtman JF, Ku GY, et al. Genetic predictors of response to systemic therapy in esophagogastric cancer. Cancer Discov. 2018;8:49–58. doi: 10.1158/2159-8290.CD-17-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute; Brown University et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- 148.Sahai E, Alberts AS, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 1998;17:1350–1361. doi: 10.1093/emboj/17.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cheng J, Demeulemeester J, Wedge DC, Vollan HKM, Pitt JJ, Russnes HG, et al. Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors. Nat Commun. 2017;8:1221. doi: 10.1038/s41467-017-01355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Biname F, Bidaud-Meynard A, Magnan L, Piquet L, Montibus B, Chabadel A, et al. Cancer-associated mutations in the protrusion-targeting region of p190RhoGAP impact tumor cell migration. J Cell Biol. 2016;214:859–873. doi: 10.1083/jcb.201601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rokutan H, Hosoda F, Hama N, Nakamura H, Totoki Y, Furukawa E, et al. Comprehensive mutation profiling of mucinous gastric carcinoma. J Pathol. 2016;240:137–148. doi: 10.1002/path.4761. [DOI] [PubMed] [Google Scholar]
- 152.Kas SM, de Ruiter JR, Schipper K, Annunziato S, Schut E, Klarenbeek S, et al. Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat Genet. 2017;49:1219–1230. doi: 10.1038/ng.3905. [DOI] [PubMed] [Google Scholar]
- 153.Cortes JR, Ambesi-Impiombato A, Couronne L, Quinn SA, Kim CS, da Silva Almeida AC, et al. RHOA g17V induces t follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018;33:259–273.:e7. doi: 10.1016/j.ccell.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zang S, Li J, Yang H, Zeng H, Han W, Zhang J, et al. Mutations in 5-methylcytosine oxidase TET2 and RhoA cooperatively disrupt T cell homeostasis. J Clin Invest. 2017;127:2998–3012. doi: 10.1172/JCI92026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chew TW, Liu XJ, Liu L, Spitsbergen JM, Gong Z, Low BC. Crosstalk of Ras and Rho: activation of RhoA abates KRas-induced liver tumorigenesis in transgenic zebrafish models. Oncogene. 2014;33:2717–2727. doi: 10.1038/onc.2013.240. [DOI] [PubMed] [Google Scholar]
- 156.Garcia-Mariscal A, Li H, Pedersen E, Peyrollier K, Ryan KM, Stanley A, et al. Loss of RhoA promotes skin tumor formation and invasion by upregulation of RhoB. Oncogene. 2018;37:847–860. doi: 10.1038/onc.2017.333. [DOI] [PubMed] [Google Scholar]
- 157.Conti MA, Saleh AD, Brinster LR, Cheng H, Chen Z, Cornelius S, et al. Conditional deletion of nonmuscle myosin II-A in mouse tongue epithelium results in squamous cell carcinoma. Sci Rep. 2015;5:14068. doi: 10.1038/srep14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liu AX, Rane N, Liu JP, Prendergast GC. Rhob is dispensable for mouse development, but it modifies susceptibility to tumor formation as well as cell adhesion and growth factor signaling in transformed cells. Mol Cell Biol. 2001;21:6906–6912. doi: 10.1128/MCB.21.20.6906-6912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fujisawa M, Sakata-Yanagimoto M, Nishizawa S, Komori D, Gershon P, Kiryu M, et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia. 2018;32:694–702. doi: 10.1038/leu.2017.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.van Hengel J, D’Hooge P, Hooghe B, Wu X, Libbrecht L, De Vos R, et al. Continuous cell injury promotes hepatic tumorigenesis in cdc42-deficient mouse liver. Gastroenterology. 2008;134:781–792. doi: 10.1053/j.gastro.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 163.Cullis J, Meiri D, Sandi MJ, Radulovich N, Kent OA, Medrano M, et al. The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. Cancer Cell. 2014;25:181–195. doi: 10.1016/j.ccr.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 164.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Arash E, Song KM, Song S, Shiban A, Attisano L. Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 2014;33:2997–3011. doi: 10.15252/embj.201490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kuhne MR, Ku G, Weiss A. A guanine nucleotide exchange factor-independent function of Vav1 in transcriptional activation. J Biol Chem. 2000;275:2185–2190. doi: 10.1074/jbc.275.3.2185. [DOI] [PubMed] [Google Scholar]
- 167.Lyons LS, Burnstein KL. Vav3, a Rho GTPase guanine nucleotide exchange factor, increases during progression to androgen independence in prostate cancer cells and potentiates androgen receptor transcriptional activity. Mol Endocrinol. 2006;20:1061–1072. doi: 10.1210/me.2005-0346. [DOI] [PubMed] [Google Scholar]
- 168.Rao S, Lyons LS, Fahrenholtz CD, Wu F, Farooq A, Balkan W, et al. A novel nuclear role for the Vav3 nucleotide exchange factor in androgen receptor coactivation in prostate cancer. Oncogene. 2012;31:716–727. doi: 10.1038/onc.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Saci A, Cantley LC, Carpenter CL. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Khanna N, Fang Y, Yoon MS, Chen J. XPLN is an endogenous inhibitor of mTORC2. Proc Natl Acad Sci USA. 2013;110:15979–15984. doi: 10.1073/pnas.1310434110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Robles-Valero J, Lorenzo-Martin LF, Menacho-Marquez M, Fernandez-Pisonero I, Abad A, Camos M, et al. A Paradoxical tumor-suppressor role for the Rac1 exchange factor Vav1 in T cell acute lymphoblastic leukemia. Cancer Cell. 2017;32:608–623.:e9. doi: 10.1016/j.ccell.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Robles-Valero J, Lorenzo-Martin LF, Fernandez-Pisonero I, Bustelo XR. Rho guanosine nucleotide exchange factors are not such bad guys after all in cancer. Small GTPases. 2018 doi: 10.1080/21541248.2018.1423851. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rack PG, Ni J, Payumo AY, Nguyen V, Crapster JA, Hovestadt V, et al. Arhgap36-dependent activation of Gli transcription factors. Proc Natl Acad Sci USA. 2014;111:11061–11066. doi: 10.1073/pnas.1322362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sauzeau V, Sevilla MA, Montero MJ, Bustelo XR. The Rho/Rac exchange factor Vav2 controls nitric oxide-dependent responses in mouse vascular smooth muscle cells. J Clin Invest. 2010;120:315–330. doi: 10.1172/JCI38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Higuchi M, Onishi K, Kikuchi C, Gotoh Y. Scaffolding function of PAK in the PDK1-Akt pathway. Nat Cell Biol. 2008;10:1356–1364. doi: 10.1038/ncb1795. [DOI] [PubMed] [Google Scholar]
- 176.Hordijk PL, ten Klooster JP, van der Kammen RA, Michiels F, Oomen LC, Collard JG. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 177.Uhlenbrock K, Eberth A, Herbrand U, Daryab N, Stege P, Meier F, et al. The RacGEF Tiam1 inhibits migration and invasion of metastatic melanoma via a novel adhesive mechanism. J Cell Sci. 2004;117(Pt 20):4863–4871. doi: 10.1242/jcs.01367. [DOI] [PubMed] [Google Scholar]
- 178.Lin Y, Zheng Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert Opin Drug Discov. 2015;10:991–1010. doi: 10.1517/17460441.2015.1058775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shang X, Marchioni F, Evelyn CR, Sipes N, Zhou X, Seibel W, et al. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci USA. 2013;110:3155–3160. doi: 10.1073/pnas.1212324110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bouquier N, Fromont S, Zeeh JC, Auziol C, Larrousse P, Robert B, et al. Aptamer-derived peptides as potent inhibitors of the oncogenic RhoGEF Tgat. Chem Biol. 2009;16:391–400. doi: 10.1016/j.chembiol.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 181.Niebel B, Wosnitza CI, Famulok M. RNA-aptamers that modulate the RhoGEF activity of Tiam1. Bioorg Med Chem. 2013;21:6239–6246. doi: 10.1016/j.bmc.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 182.Wang J, Hu K, Guo J, Cheng F, Lv J, Jiang W, et al. Suppression of KRas-mutant cancer through the combined inhibition of KRAS with PLK1 and ROCK. Nat Commun. 2016;7:11363. doi: 10.1038/ncomms11363. [DOI] [PMC free article] [PubMed] [Google Scholar]