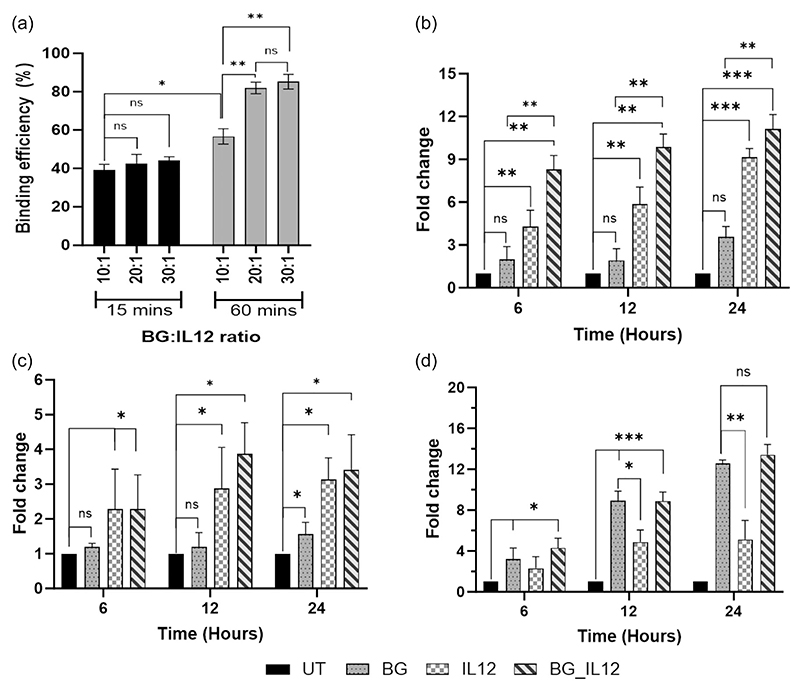
Figure 4. Interleukin-12 binding and gene expression analyses.
(a) Quantification of interleukin-12 (IL12) binding with BGs was assessed for varying BG:IL12 ratios and coincubation periods using a sandwich enzyme-linked immunosorbent assay (ELISA). The graph depicts the percentage of IL12 binding, calculated relative to the initial amount of IL12 used in the experiment. Relative quantification of gene expression was performed via qPCR on BG/IL12/BG_IL12-treated NK92 cells using the 2–ΔΔCt method to determine the fold change in the expression of perforin (b), granzyme-B (c), and Fas-L (d), relative to the untreated control. GAPDH was used for normalization. *p < 0.05, **p < 0.005, and ***p < 0.001, ns: not significant. BG, bacterial ghost; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.