Fig. 3. Analysis of 367 complex crystal structures reveals hotspots for ligand engagement and a variety of ways to engage substrate recognition subpockets.
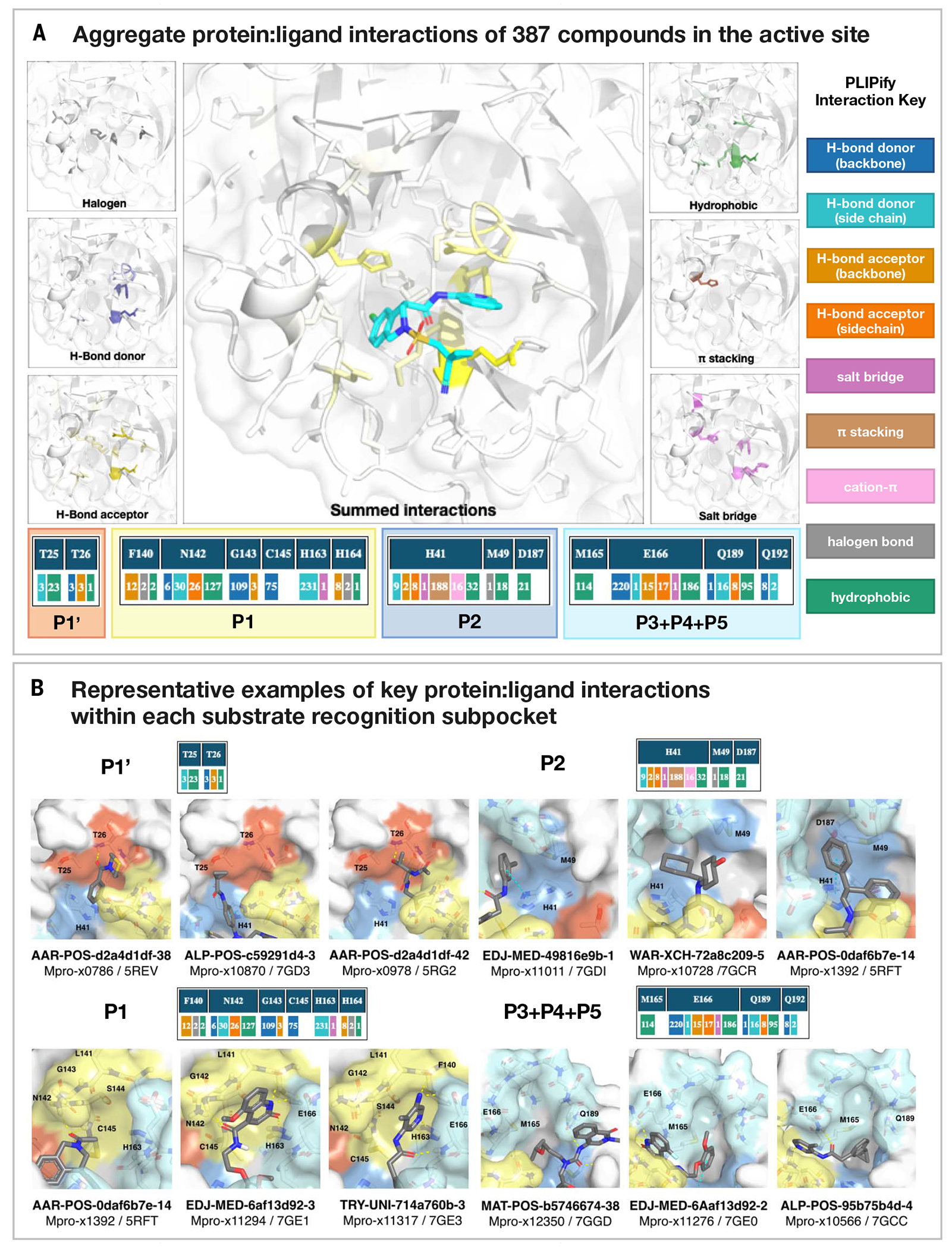
(A) The five substrate recognition subpockets exhibit distinct preferences for intermolecular interactions. The figure highlights the locations of different types of interactions, with the shading indicating the frequency. The bottom row tallies the number of times that each interaction was seen in our structures for different residues. The interaction map was generated using PLIPify (Materials and methods) and summarizes the interactions witnessed across 367 complexes from the perspective of the protein, distinguishing between backbone (bb) and sidechain (sc) interactions (which might be more vulnerable to point mutations). (B) Representative examples of protein-ligand interactions engaging the P1’, P1, P2, and P3-5 subpockets. Hydrogen bonds and π-stacking interactions are depicted as yellow and cyan dashed lines, respectively. The rows above each set of complexes tally the number of times that each interaction was seen with the specific residues within the subpockets. See data S4 for Protein Data Bank (PDB) IDs and crystallography statistics. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
