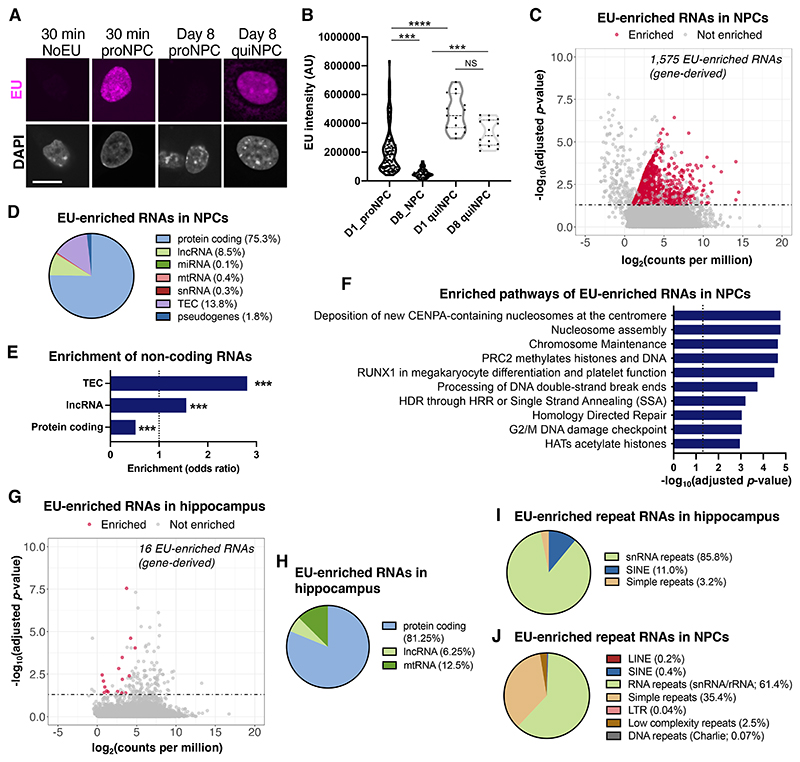
Fig. 2. Molecular identity of long-retained RNA.
(A) Retention of EU signals in quiescent NPCs (quiNPCs) in vitro. Proliferating NPC (proNPC). Scale bar, 10 μm. (B) Quantification of EU intensity in proNPCs and quiNPCs at day 1 (D1) or day 8 (D8) after EU labeling. Kruskal-Wallis test followed by Dunn’s multiple comparison test; ***P < 0.001, ****P < 0.0001, dots indicate individual cells. (C) MA plot showing EU-enriched (gene-derived) RNAs in quiNPCs at 8 days after EU labeling, as determined by EU-RNA-Sequencing. Significantly EU-enriched RNAs were defined as adjusted P < 0.05 and fold change > 2 compared to non-EU-labeled quiNPCs (n=3 experiments). (D) Gene class distribution of EU-enriched RNA in quiNPCs. (E) Protein-coding RNAs were underrepresented, whereas long non-coding RNAs (lncRNA) and uncharacterized transcripts (TEC) were over-represented in EU-enriched RNA. ***P < 0.001 (linear regression). (F) Top significantly enriched Reactome pathways among EU-enriched RNAs in quiNPCs (adjusted P < 0.05). (G) MA plot showing EU-enriched (gene-derived) RNAs in hippocampus tissue of 5-week-old mice that were injected with EU at postnatal days P3-P5 (n = 3 mice/group). (H) Gene class distribution of EU-enriched RNA in the hippocampus. (I-J) Distribution of EU-enriched RNAs that map to repeat RNAs in quiNPCs and hippocampus tissue.