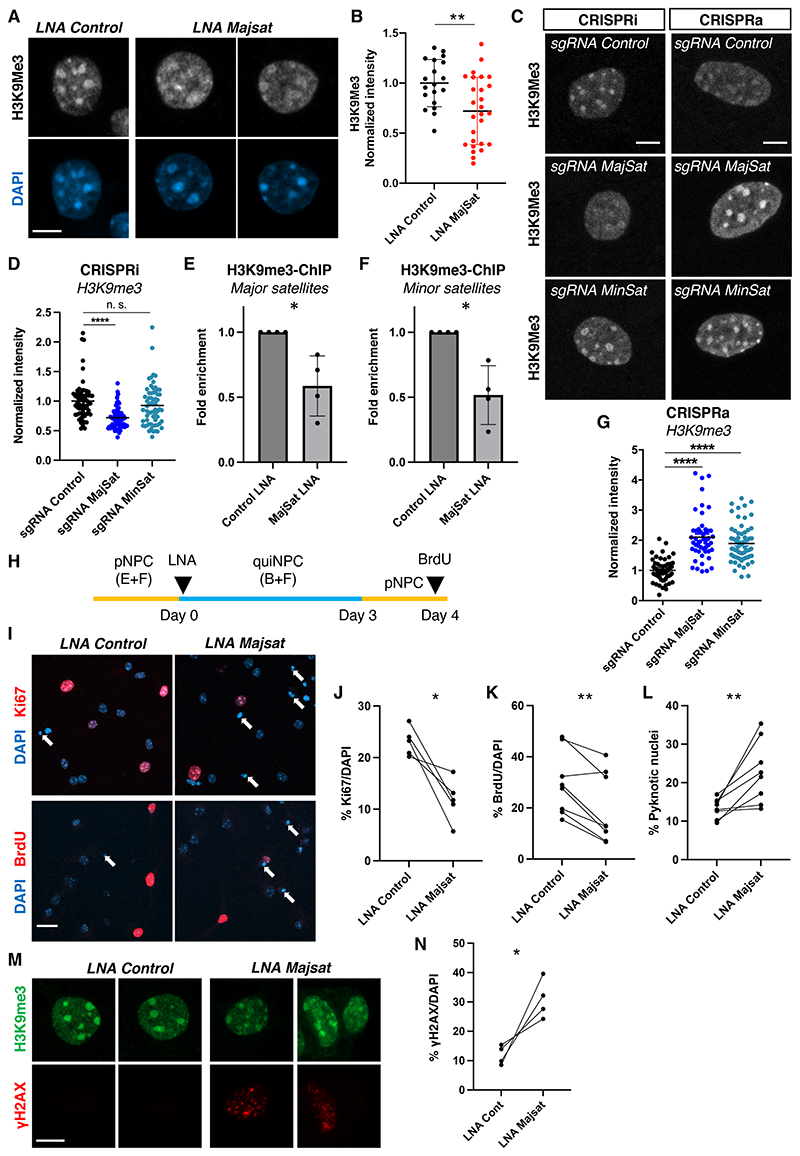
Fig. 4. Major satRNAs are essential for maintenance of NPC function.
(A) Distribution of H3K9me3 after LNA-mediated knock-down of major satRNAs in quiNPCs. Scale bar, 5 μm. (B) Quantification of H3K9me3 intensity after knock-down of major satRNAs. **P < 0.01, t-test, dots indicate individual cells from 3 experiments. (C) Distribution of H3K9me3 after CRISPR-mediated transcriptional inhibition (CRISPRi) or overexpression (CRISPRa) of major and minor satRNAs. Scale bar, 5 μm. (D) Transcriptional inhibition of major satRNAs using CRISPRi (dCas9-KRAB with sgRNAs) impaired heterochromatin retention in quiNPCs; minor satRNAs were dispensable. ***P < 0.001, n. s. not significant, t-test, dots represent individual nuclei from 3 independent experiments, group means are indicated. (E-F) LNA-mediated knock-down of major satRNAs reduced H3K9me3 abundance at major and minor satellite repeats. Shown are data from H3K9me3-ChIP-qPCR (*P < 0.05; one-sided t-test, n = 4). Data are presented as mean +/- SD. (G) Upregulation of satRNAs increased nuclear H3K9me3 in quiNPCs. ****P < 0.0001, t-test, dots indicate individual cells from 3 independent experiments, group means are indicated. (H) Experimental timeline for data shown in (I-N). (I) Cell cycle re-entry of quiNPCs after LNA-mediated knock-down of major satRNAs. Arrows indicate pyknotic cells. Scale bar, 25 μm. (J-K) Decreased percentage of Ki67+ and BrdU+ cells one day after re-activation of quiNPCs into proliferation. *P < 0.05, **P < 0.01, paired t-test, n = 5-8. Ki67, LNA control 23.1 ± 2.73%, LNA Majsat 11.8 ± 4.16%; BrdU, LNA control 29.6 ±12.35%, LNA Majsat 19.65 ± 13.64%. (L) Increased percentage of pyknotic cells after knock-down of major satRNAs. LNA control 13.4 ± 2.55%, LNA Majsat 22.76 ± 8.11%. (M-N) Increased percentage of γH2AX+ cells after knock-down of major satRNAs. *P < 0.05, ratio paired t-test, n = 4. LNA control 8.57 ± 3.25%, LNA Majsat 30.9 ± 6.66%. Scale bar in M, 5 μm.