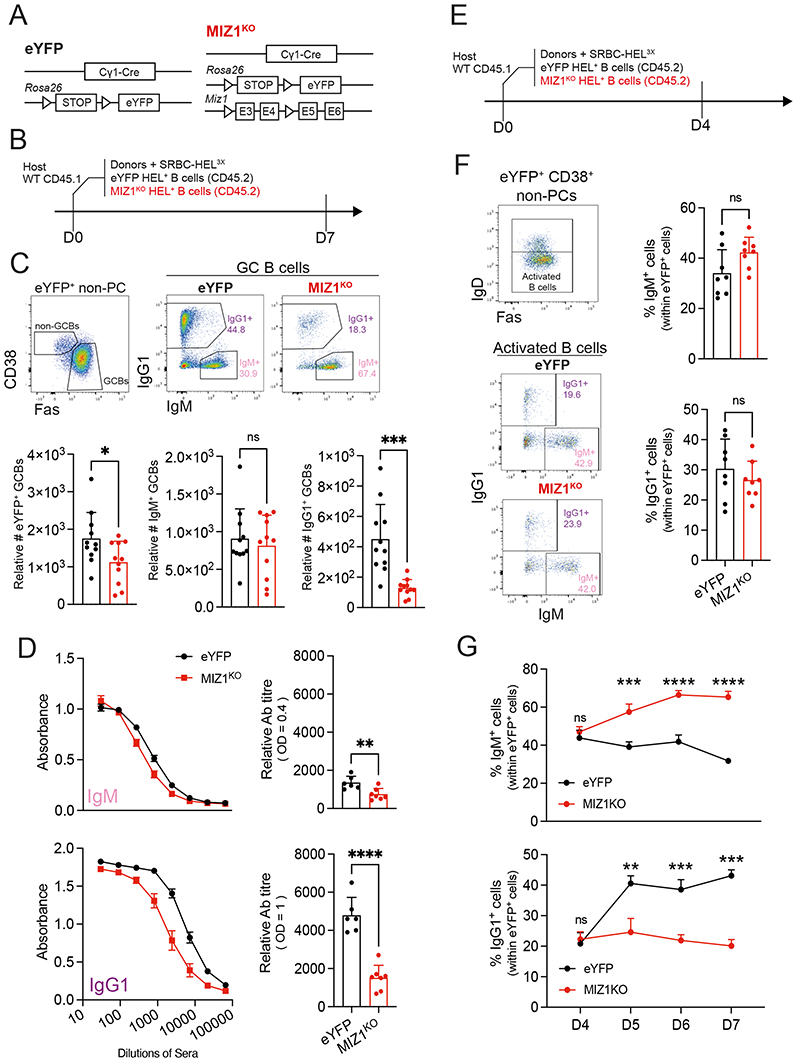
Fig. 2. Miz1 loss impairs IgG1+ GC B cell accumulation in vivo.
(A) Schematics of allele combination in control (eYFP) and experimental mice (MIZ1KO). An allele containing loxP-stop-loxP-eYFP (loxP= triangle) in the Rosa26 locus is present in both eYFP and MIZ1KO mice. MIZ1KO contains an Miz1 allele with two loxP (triangle) flanking exon3 and exon4 that encode the BTB/POZ DNA binding domain.
(B) SWHEL experiment schematics.
(C) Top, gating strategy for IgG1+ and IgM+ GC B cell subsets generated from donor-derived reporter positive B cells. Bottom, graphs displaying cumulative data of relative cell numbers (numbers per 106 splenocytes) for total GC B cells, and GC B cell IgG1+ and IgM+ subsets in, eYFP (control, black); and MIZ1KO (red).
(D) Left, ELISA for HEL-binding IgG1 and IgM antibodies in recipient mice. Right, relative antibody titer of anti-HEL IgG1 (OD = 1), and anti-HEL IgM (OD = 0.4).
(E) SWHEL experiment schematics as in (B). Recipient mice spleen and sera were analyzed at day 4 (F) after immunization.
(F) Left, gating strategy; right, percentages of IgM+ and IgG1+ subsets within early activated B cells.
(G) Dynamic changes of the percentages of IgG1+ and IgM+ B cell subsets in activated B cells (D4), and GC B cells (D5-7).
Each symbol (C: eYFP n = 12, MIZ1KO n = 12; D : eYFP n = 6, MIZ1KO n = 7; F: day 4 eYFP n = 8, MIZ1KO n = 8; H: day 4 eYFP n = 3, MIZ1KO n = 3; day 5, 6 eYFP n = 6, MIZ1KO n = 6; day 7 eYFP n = 3, MIZ1KO n = 3) represents an individual mouse; small horizontal lines show mean and SEM. Data in (C-D) is representative of three to five experiments. Data in (F) is from two independent experiments. Data in (G), D4 and D7 is data from one experiment, D5-6 is data from two independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 (unpaired two-tailed Student’s t test in (C-D, F); multiple T tests in (G)). ns, not significant.