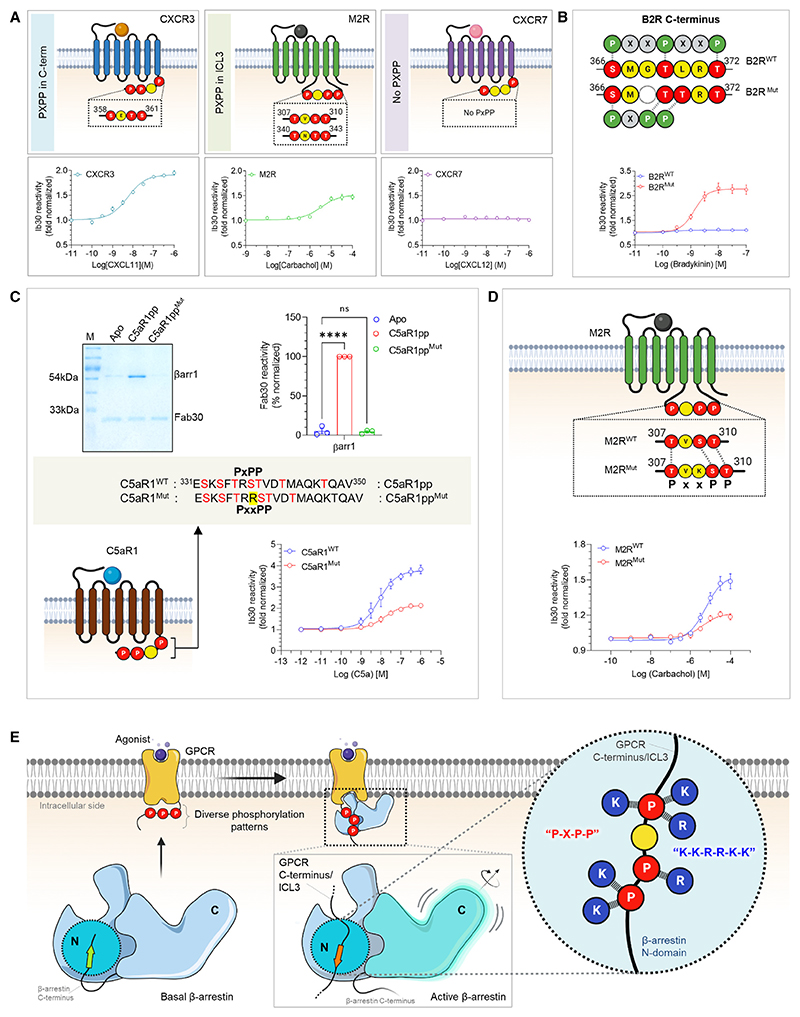
Figure 7. A key phosphorylation motif for βarr activation.
(A) NanoBiT-based assay for assessing Ib30 reactivity to CXCR3 (left), M2R (middle), and CXCR7 (right) activated βarr1 (Receptor+SmBiT-βarr1+LgBiT-Ib30) (mean ± SEM; n = 3; normalized as fold over basal).
(B) Deletion of G368 and substitution of L370 to Thr in B2R engineers the P-X-P-P (referred to as B2RMut) and results in gain of function in terms of Ib30 reactivity as measured using the NanoBiT assay (Receptor+SmBiT-βarr1+LgBiT-Ib30) (mean ± SEM; n = 3; normalized as fold over basal).
(C) Addition of an extra Arg between positions 336 and 337 in C5aR1pp to disrupt the P-X-P-P (referred to as C5aR1ppMut) leads to a near-complete loss of Fab30 (top) reactivity as measured in coIP assay (mean ± SEM; n = 3; densitometry-based data normalized with respect to C5aR1pp signal as 100%; one-way ANOVA, Dunnett’s multiple comparisons test). The exact p values are as follows: Apo vs. C5aR1pp2 (p < 0.0001), Apo vs. C5aR1pp4 (p = 0.9160) (****p < 0.0001, ns, non-significant). Corresponding mutation in C5aR1 to disrupt the P-X-P-P (referred to as C5aR1Mut) results in a dramatic decrease in Ib30 reactivity (bottom) as measured using the NanoBiT assay (mean ± SEM; n = 5; normalized as fold over basal).
(D) Insertion of a Lys residue between positions 308 and 309 to disrupt the P-X-P-P pattern in ICL3 of M2R (referred to as M2RMut) shows reduced Ib30 reactivity compared with the wild type (M2RWT) as measured using the NanoBiT assay (mean ± SEM; n = 4; normalized as fold over basal).
(E) Schematic representation summarizing the identification of a phosphorylation motif in GPCRs that drives βarr activation. See also Figure S6.