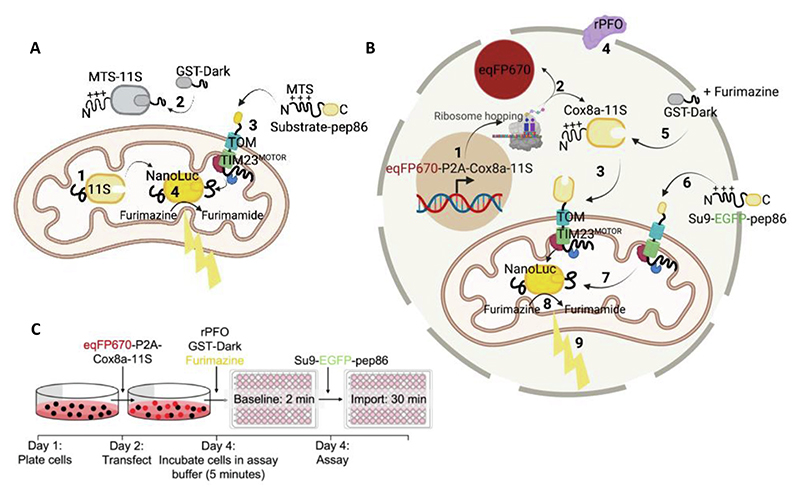
Figure 1. The MitoLuc Assay System for Monitoring Mitochondrial Protein Import in Yeast Mitochondria and Permeabilised Cells.
(A) Schematic showing concept of the MitoLuc system to monitor mitochondrial import in isolated yeast mitochondria. Mitochondria were isolated from yeast producing a fusion of the mitochondrial targeting presequence (MTS) and the large fragment of split MitoLuc, 11S (MTS-11S). The resultant preparation with matrix localised 11S (with cleaved MTS) was deployed in the import assay: (1). GST-Dark is added to bind non-mitochondrial 11S, decreasing background signal (2). A pep86-tagged substrate protein (containing an N-terminal MTS; Su9-EGFP-pep86) is added and targeted to the mitochondrial matrix (3). The MTS is cleaved and pep86 binds to 11S in the matrix, forming MitoLuc (4). This converts furimazine to furimamide producing a luminescent signal. Schematic created using BioRender. (B) Schematic showing the concept of the permeabilised MitoLuc import assay system in live mammalian cells. DNA encoding eqFP670 (far red fluorophore) followed by a P2A region and Cox8a-11S is transcribed in the nucleus and translated in the cytosol (1), leading to the production of cytosolic eqFP670 and mitochondrially targeted Cox8a-11S (2). Cox8a-11S is translocated to the mitochondrial matrix via the presequence pathway (3). At the time of the assay, MitoLuc assay buffer containing 3 nM rPFO is added, which perforates the plasma membrane (4) whilst retaining intact mitochondria. This allows other substrates, drugs, furimazine, and proteins to enter the cells. One such protein is GST-Dark, which mops up any remaining cytosolic 11S (5), preventing background signal from cytosolic binding. Following a baseline read, a pep86 containing precursor is added (in this case Su9-EGFP-pep86; (6)) which is translocated into mitochondria where it binds to 11S (7), forming the functional MitoLuc enzyme, which converts furimazine to furimamide (8), producing a bioluminescent signal corresponding to import (9). Schematic created using BioRender. (C) Experimental outline of the permeabilised MitoLuc assay. On day 1, cells are plated, and are subsequently transfected with eqFP670-P2A-Cox8a-11S DNA on day 2. On day 4, the day of the assay, media is replaced with assay buffer containing furimazine, rPFO, and GST-Dark and incubated for 5 minutes prior to carrying out a 2 minute baseline read, followed by injection of the Su9-EGFP-pep86 protein and a 30-minute kinetic import read for luminescence corresponding to protein import.