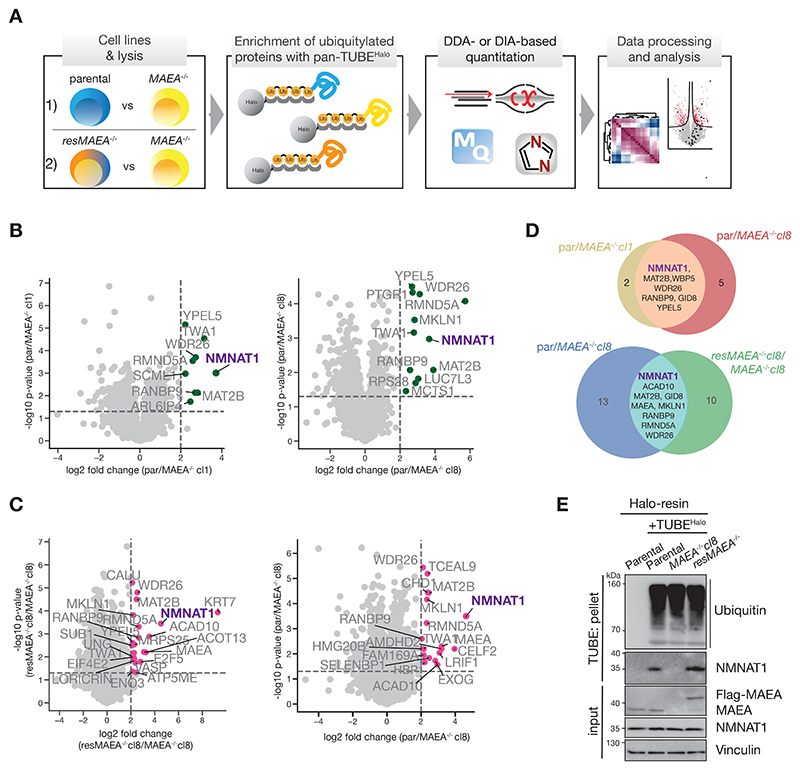
Figure 1. Ubiquitome-enrichment proteomics reveals a CTLH E3 substrate.
A) Workflow of the proteomics study indicating the used HEK293 cell systems (1) parental vs MAEA-/- knockout and (2) MAEA-/- + MAEA rescue line (resMAEA-/-) vs MAEA-/-, enrichment of ubiquitylated proteins with Halo-tagged pan-TUBE (TUBEHalo), followed by DDA- or DIA-quantitation and data processing/analysis.
B) Individual volcano plots of the -log10 p-values vs the log2 protein abundance of TUBE-precipitates between parental and two MAEA-/- clones (cl1 and cl8). 5% p-value and 4x cut off are indicated.
C) Individual volcano plots of the -log10 p-values vs the log2 protein abundance of TUBE-precipitates between resMAEA-/- vs MAEA-/-cl8 and between parental vs MAEA-/- cl8. 5% p-value and 4x cut off are indicated.
D) Venn diagrams showing overlapping proteins from 5% p-value and 4x cut off of B and C.
E) TUBEHalo-captured proteins from MAEA-/-cl8 and resMAEA-/- cell lines were precipitated with Halo-resin (TUBE: pellet) and analyzed by immunoblot analysis. Vinculin serves as protein input control.