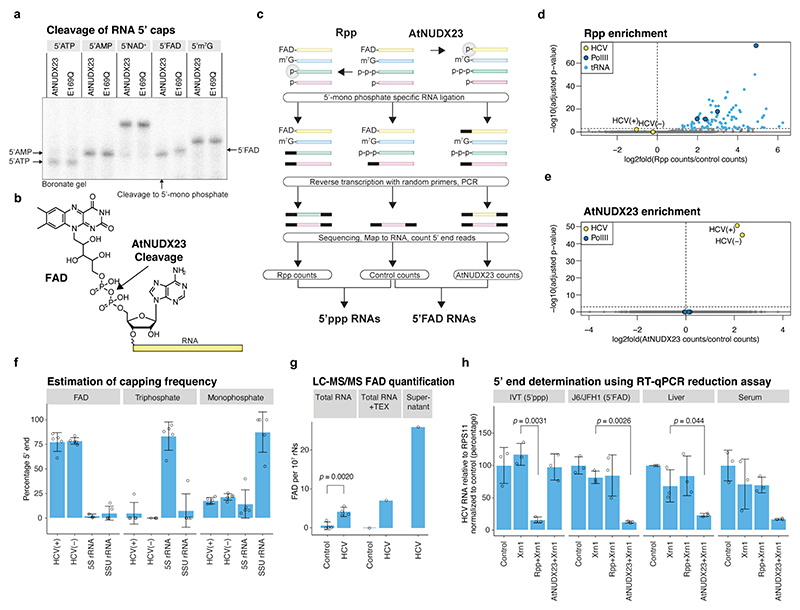
Fig. 1 |. Demonstration of HCV FAD capping.
(a) AtNUDX23 specificity against different 5’caps on a 40 nt RNA. Reactions were analysed on a boronate gel, which affects the migration of RNAs depending on their 5’ cap. E169Q is a catalytically inactive AtNUDX23 mutant. (b) AtNUDX23 cleavage of FAD-capped RNA. (c) Schematic representation of the CapZyme-seq method. (d) Identification of 5’ppp RNAs from J6/JFH1 infected Huh7.5 cells. Volcano plot showing DeSeq2 adjusted p-values versus log2(fold change) for reads mapping to the 5’ termini of individual RNAs (n=5 biological replicates). Rpp treated samples are compared to control samples. (e) Identification of 5’FAD capped RNAs. As in (d), but AtNUDX23 treated samples are compared to control samples. (f) Estimation of the percentage of FAD, tri- and mono-phosphate 5’ termini for J6/JFH1(+) and (−) RNA, 5S rRNA and SSU rRNA (n=5 biological replicates). The analysis assumes stoichiometric conversion to monophosphate by Rpp and AtNUDX23. (g) LC-MS/MS FAD quantification using stable isotope-labelled internal standards for FAD and normalization to ribonucleotide content. RNA samples were isolated from J6/JFH1 infected and control Huh7.5 cells (n=4 biological replicates) and single samples of TEX treated J6/JFH1 infected and control Huh7.5 cells plus ultrafiltration supernatant from HCV infected Huh7.5 cells. TEX: Terminator 5’ exonuclease. (h) RT-qPCR reduction assay showing HCV RNA levels after treatment with Xrn1 5’ exonuclease (5’p specific reduction), Rpp + Xrn1 (5’ppp reduction) or AtNUDX23 + Xrn1 (5’FAD reduction). The following samples were compared: in vitro transcribed 5’ppp J6/JFH1 (IVT), RNA extracted from J6/JFH1 infected Huh7.5 cells (J6/JFH1), and liver and plasma from a J6/JFH1A876P infected human liver chimeric uPA-SCID mouse (n=3 independent replicates). The p-values in (g) and (h) are calculated using one-sided Welch’s unequal variances t-test. Data are presented as mean +/− SD. For gel source data, see Supplementary Figure 1.