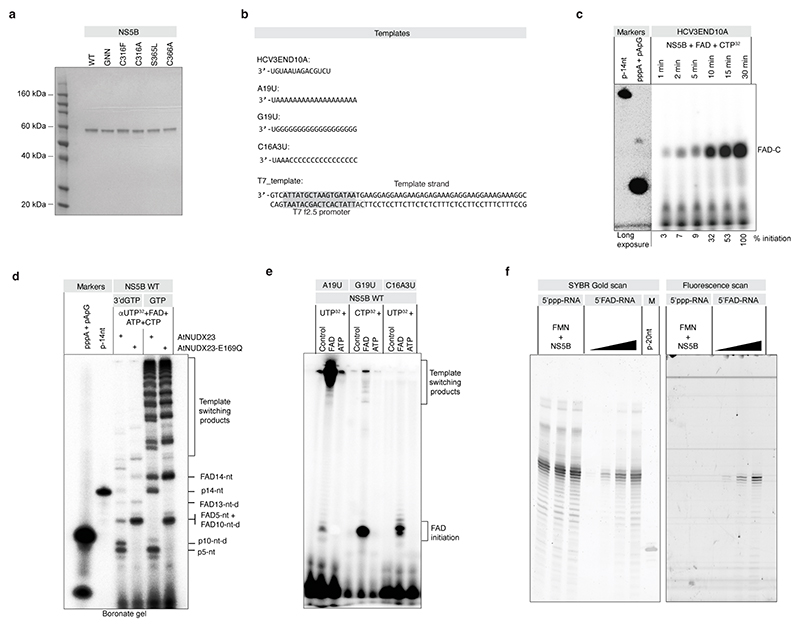
Extended Data Fig.7 |. FAD dependent replication initiation by HCV NS5B.
(a) Coomassie stained SDS-PAGE of the HCV NS5B WT, GNN, C316F, C316A, S365L and C366A recombinant proteins. (b) RNA templates used for in vitro replication reactions. (c) Time-dependent formation of the initiation product. HCV NS5B polymerase was incubated with HCV3END10A RNA in the presence of FAD and CTP, complemented with α 32P-labeled CTP for visualization. Reactions were terminated at the indicated time points. Products were resolved with 18 % denaturing PAGE together with 5’ radioactively labelled markers. (d) The replication extension products contain FAD. NS5B polymerase was incubated with HCV3END10A RNA in the presence of FAD, ATP, CTP, UTP (complemented with α-32P-labeled UTP) and either GTP or 3’dGTP. After 1 hr incubation, the reactions were treated with AtNUDX23 or AtNUDX23-E169Q. The products were resolved with 18 % denaturing 0.2 % boronate PAGE. The replication reactions subjected to AtNUDX23 or AtNUDX23-E169Q treatment are identical to the α-32P-labeled UTP reactions separated on a regular PAGE gel in main Fig. 3c. The presence of boronate retards FAD-capped RNAs compared to uncapped RNAs due to the relatively stable diol complexes formed between the gel derived boronyl groups and naturally present diols of the RNA 3’ end and of FAD. Accordingly, both 5’ FAD capping and 3’ deoxy termination of extension products will affect migration in the boronate gel. Products were annotated by comparison between the 3’dGTP/GTP conditions, AtNUDX23/AtNUDX23-E169Q conditions and the migration observed on the regular PAGE gel (main Fig. 3c). (e) FAD promotes replication initiation on different RNA templates. Indicated RNA templates were incubated with HCV NS5B polymerase. ATP, FAD or control was added as initiating nucleotide. Elongating UTP or CTP nucleotides were supplemented with α32P-labeled UTP or CTP, respectively. The resulting products were resolved with 18 % denaturing PAGE. (f) Lack of HCV NS5B mediated post-initiation FAD capping. To exclude that NS5B mediated 5’FAD capping could happen post-initiation on a 5’ppp initiated RNA, 5’ppp RNA was incubated with FMN in the presence of NS5B polymerase. RNA was resolved with 18 % PAGE and visualised by nucleic acid staining (left) and with fluorescent signal indicative of the 5’FAD cap (right). Increasing concentrations of 5’FAD capped RNA generated with T7 polymerase were used as markers on the same gel. For gel source data, see Supplementary Figure 1.