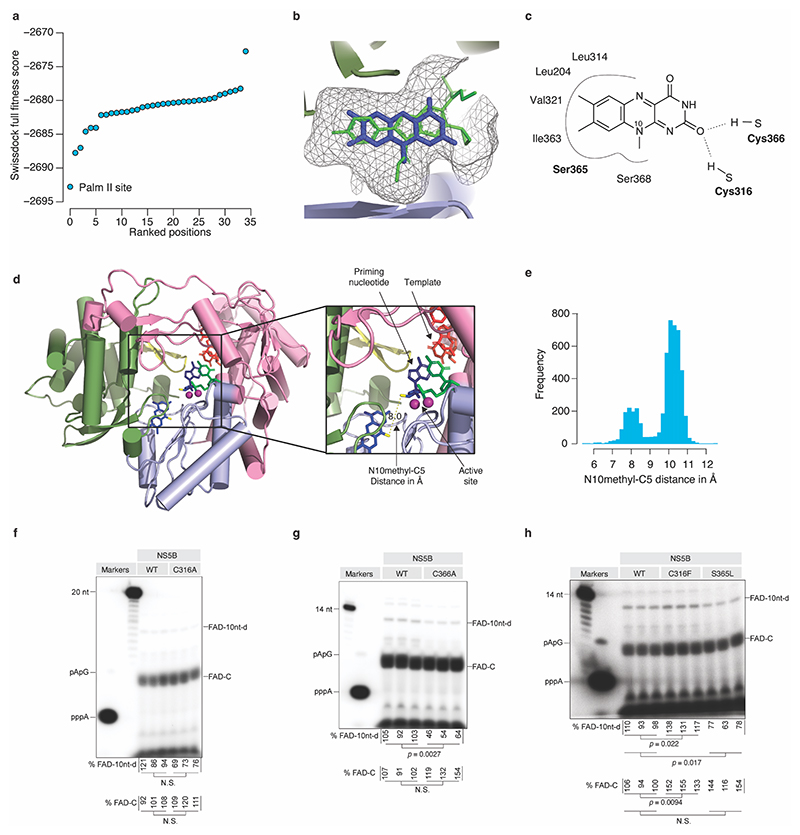
Extended Data Fig.8 |. Prediction model of FAD − HCV NS5B interaction.
(a) In silico docking analysis of FAD into HCV NS5B using SwissDock. Full fitness scores for the top 36 positions obtained by docking of lumiflavin into HCV NS5B are shown. (b) Close-up view of the palm II binding pocket demonstrating the overlap between the nesbuvir (green) binding site and the putative binding site of the lumiflavin moiety of FAD (blue). The docking was based on (PDB: 3FQL) with the nesbuvir molecule removed. (c) Predicted contacts between NS5B and lumiflavin when docked in the palm II site of NS5B. (d) Predicted lumiflavin binding site modelled into the structure of the NS5B primed initiation complex (PDB:4WTM) colored by subdomains: fingers (pink), palm (light blue), thumb (pale green) and beta-loop (yellow). The HCV(+) 3’ end sequence was modeled into the position of the template (red). An adenosine (blue) and CMP (green) were modeled into the position of the priming and incoming nucleotides, respectively. The close-up view shows the active site with two localized Mn2+ ions in violet. The direct distance between the lumiflavin N10 methyl and the ribose C5 is indicated. (e) Observed FAD N10 methyl-C5 distances from 4,928 crystal structures of proteins containing a FAD cofactor. (f-h) Triplicate analysis (independent replicates) of de novo initiation with FAD for the indicated NS5B mutants using the HCV3END10A template with the FAD extension and initiation signal quantified below the gels. The p-values are calculated using two-sided Welch’s unequal variances t-test. N.S. equals p-val > 0.05. For gel source data, see Supplementary Figure 1.