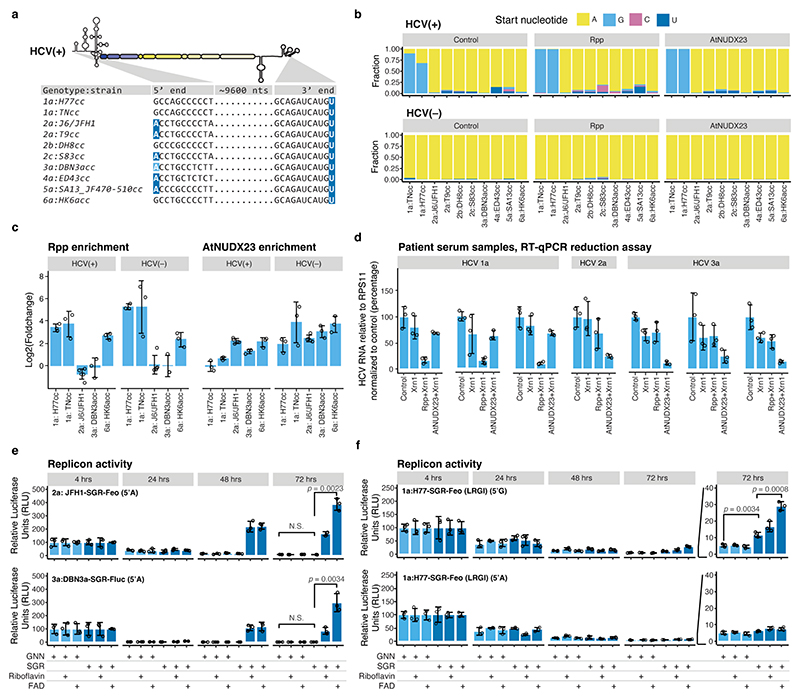
Fig. 2 |. FAD capping for HCV strains of genotypes 1-6.
(a) Alignment showing the 5’ and 3’ termini of the HCV strains used in this study. Residues compatible with FAD capping are highlighted in dark blue. Genotype 3a isolates in general, as well as DBN, have 5’A compatible with FAD capping, however, the DBN3acc clone used had a non-authentic 5’G that reverts in culture (light blue). (b) Observed 5’ nucleotides in control, Rpp and AtNUDX23 enriched CapZyme-seq libraries for the HCV(+) and (–) RNAs after propagation of the indicated strains in Huh7.5 cell culture. (c) Enrichment of 5’ terminal reads for HCV(+) and (–) RNAs. Shown are DESeq2 log2FoldChange observed in Rpp (5’ppp, left) and AtNUDX23 (5’FAD, right) enriched CapZyme-seq libraries from RNA isolated from Huh7.5 cells infected with indicated HCV strains. Error bars show the DESeq2 standard error of the log2FoldChange estimate, n=3 biological replicates, except J6/JFH1 with n=5. (d) RT-qPCR reduction assays on RNA isolated from plasma samples taken from patients infected with HCV genotypes 1-3 (technical triplicate for each sample). (e) Replication of wildtype (SGR) and corresponding non-replicating mutant (GNN) of the JFH1-SGR-Feo and DBN3a-SGR-Fluc replicons in Huh7.5 cells grown in riboflavin-depleted media shown as relative luciferase units (RLU) relative to 4 hrs at the indicated time points. (f) Replication of H77-SGR-Feo (LRGI) replicon with authentic 5’G (top) or engineered 5’A (bottom) presented as in panel (e). Transient replication levels are generally lower for H77-SGR-Feo (LRGI) than for JFH1-SGR-Feo and DBN3a-SGR-Fluc. The p-values in (d-f) are calculated using one-sided Welch’s unequal variances t-test. Data are presented as mean +/− SD (n=3 biological replicates); N.S. are non-significant differences. Riboflavin (0.4 mg/L) or FAD (10 μM) was used for reconstitution in (e-f).