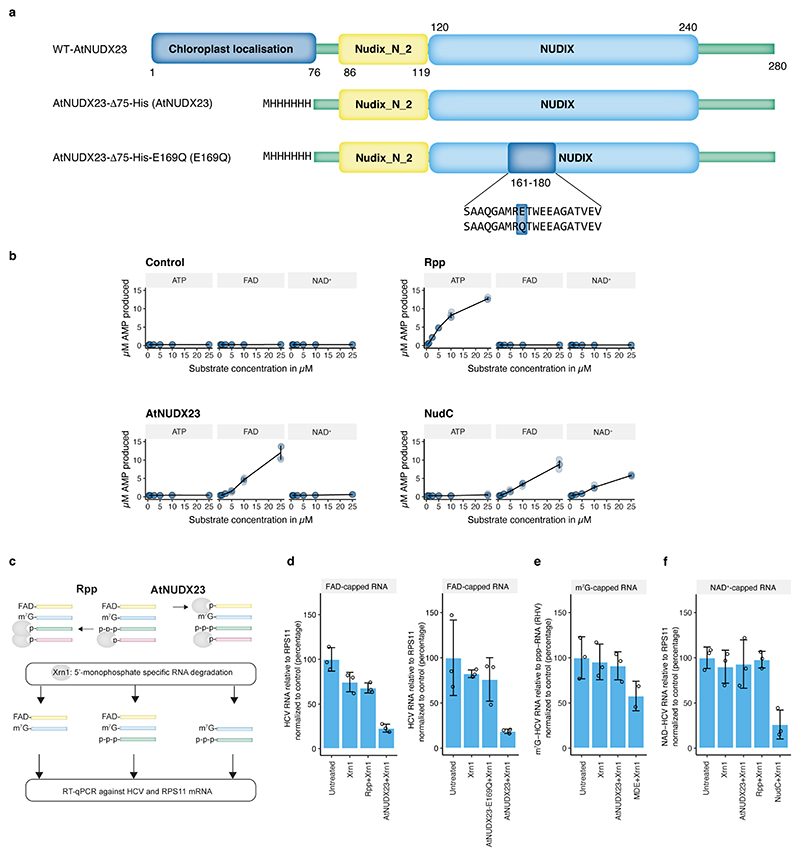
Extended Data Fig.1 |. Specificity of enzymes used for CapZyme-seq enrichment.
(a) Schematic representation of the AtNUDX23 and AtNUDX23-E169Q proteins used in this study. For both, the N-terminal 75 residues were truncated to remove the chloroplast localisation signal present in the WT protein. (b) AMP production in the presence of increasing concentrations of ATP, FAD and NAD+ for the indicated enzymes and the no enzyme control (n=4, independent replicates). Data are presented as mean +/− SD. (c) Strategy for the RT-qPCR reduction assay. (d) RT-qPCR reduction assay testing activity of AtNUDX23 and Rpp against in vitro transcribed FAD-capped RNA (left) and showing the lack of activity of the AtNUDX23-E169Q protein (right). (e) RT-qPCR reduction assay testing activity of AtNUDX23 and mRNA decapping enzyme (MDE) against in vitro transcribed m7G-capped RNA. (f) RT-qPCR reduction assay testing activity of AtNUDX23, Rpp and NudC against in vitro transcribed NAD+-capped RNA. For (d-f), data are presented as mean +/− SD, n=3 independent replicates.