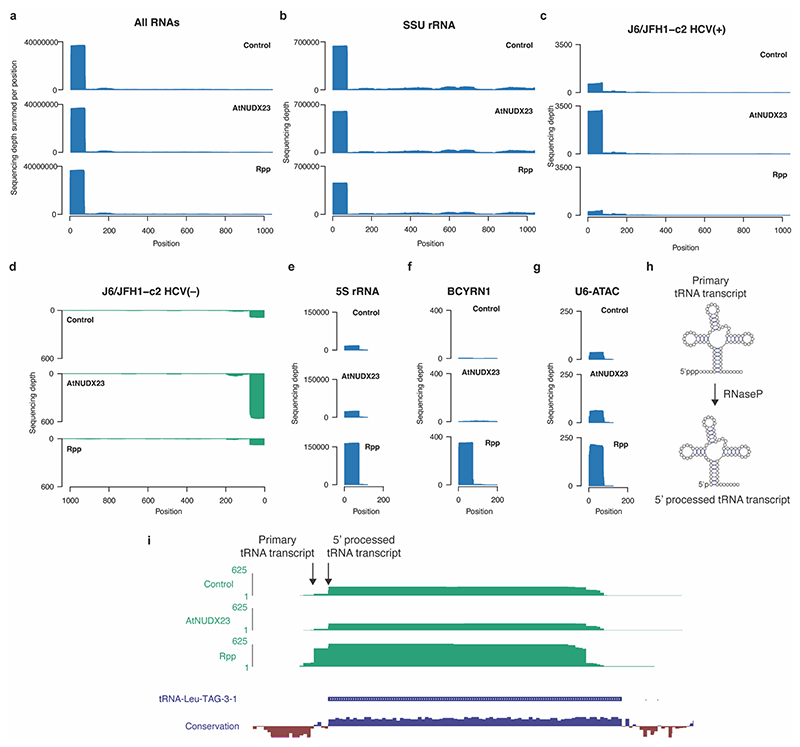
Extended Data Fig.2 |. CapZyme-seq reads map to 5’ termini of RNAs.
Normalized sequencing depth plots for CapZyme-seq libraries: no enzyme control, AtNUDX23 and Rpp enrichment. Reads were mapped to human noncoding RNAs plus the most highly expressed mRNAs (> 2 TPM) in Huh7.5 cells. The normalized sequencing depth is shown for (a) all RNAs combined, (b) SSU rRNA, (c) J6/JFH1-c2 HCV(+) RNA, (d) J6/JFH1-c2 HCV(–) RNA, (e) 5S rRNA, (f) BCYRN1 RNA and (g) U6-ATAC RNA. (h) Schematic representation of tRNA processing: RNase P mediates processing of 5’ppp termini of primary tRNAs to 5’p mature tRNAs. (i) Normalized CapZyme-seq sequencing depth for tRNA-Leu-TAG-3-1 is shown for no enzyme control, AtNUDX23- and Rpp enrichment (top panels). Base wise conservation (phyloP score) for 100 vertebrate species and gene annotation (both obtained from UCSC genome browser) are shown in the bottom panels. Conserved positions have positive phyloP scores (blue). For browsing of the CapZyme-seq data at other human genomic loci, the following UCSC genome session can be used: http://www.genome.ucsc.edu/s/vinther/hg19_CapZyme%2Dseq_tRNA%2DLeu%2DTAG