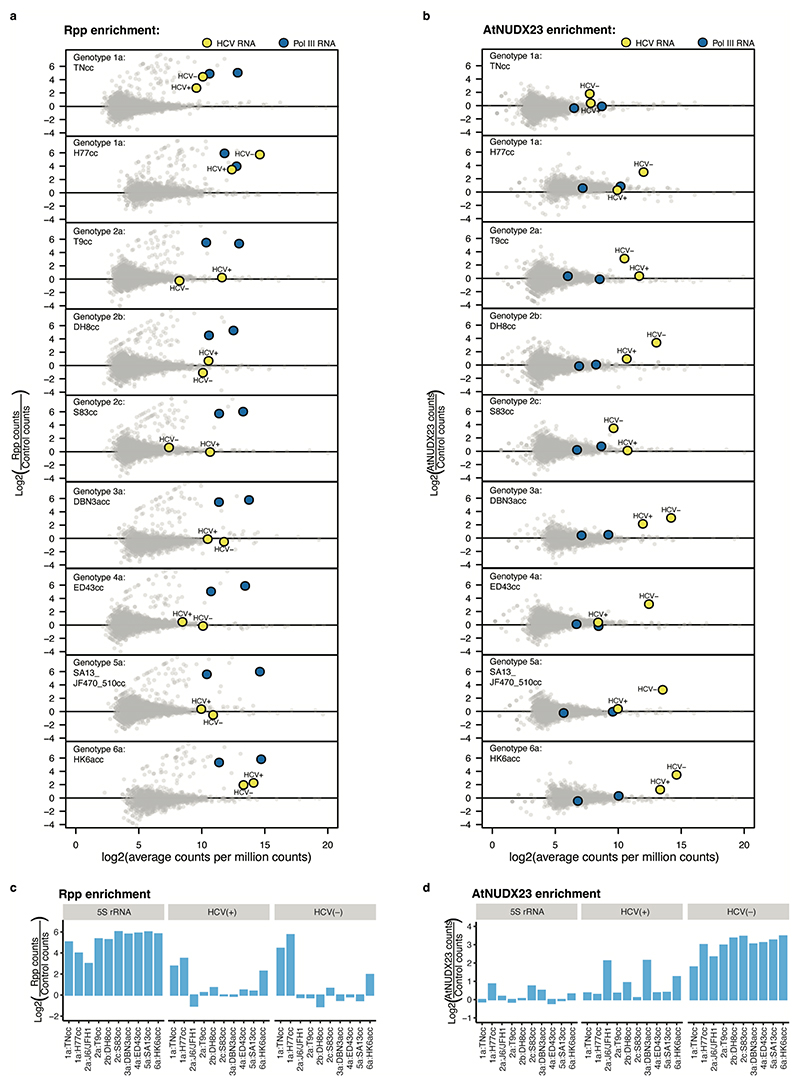
Extended Data Fig.5 |. Identification of 5’ capping for different HCV strains.
CapZyme-seq analysis of RNA isolated from Huh7.5 cells infected with the indicated HCV genotypes/strains. Mean-difference plots show log2 (fold changes) as a function of log2 (average abundance) for reads mapping to the 5’ termini of individual RNA molecules. Enrichment with Rpp (a) and AtNUDX23 (b) was based on comparison of enzyme-treated libraries to no enzyme control libraries. The log2 (fold values) observed for HCV(+), HCV(–) and 5S rRNA (5’ppp) for the different genotypes/strains are summarized in (c) for Rpp and (d) for AtNUDX23. Data is derived from a single biological replicate. Main Fig. 2c shows replicate experiments for selected strains.