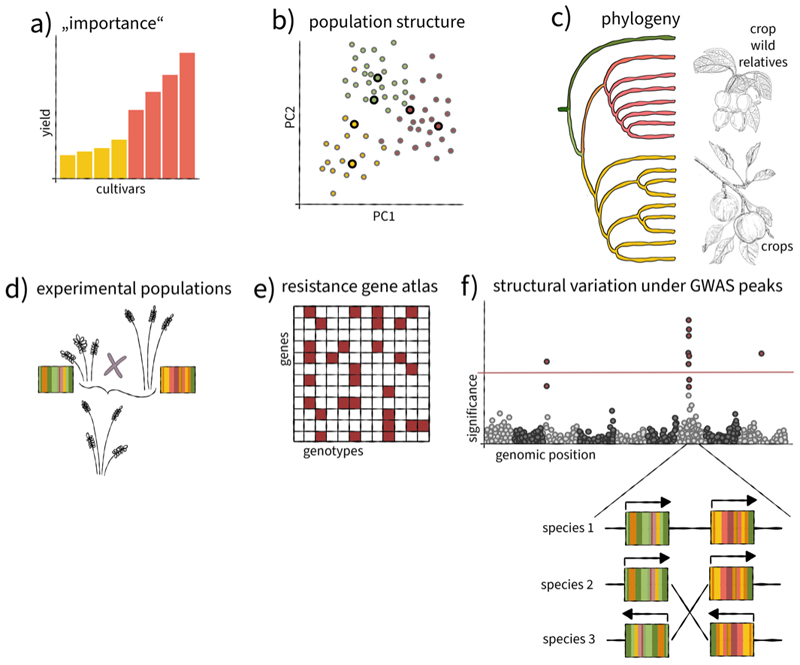
Figure 2. Pangenomics in crop plants.
Most pangenome studies to date have focused on crops. The varieties under investigation are selected based on different criteria.(a) Cultivars of great ‘importance’ include those that are widely grown or used in genetic research. (b) Surveys of population structure enable the selection of core sets that represent with a limited number of samples genetic diversity in a given crop as best as possible. The diversity space of species is often represented in principal component analysis (PCA). Population structure is reflected in clusters (shown in different colours) that correspond to geographic origins or infraspecific taxonomy. (c) Crop-wild relatives (wild progenitors and more distant relatives) are studied because they broaden allelic diversity in cultivated varieties. (d) Pangenomes have diverse applications in crop genetics. Genome sequences of the parents of experimental population assist in mapping traits to single genetic factors (coloured bar). (d). (e) Catalogues of resistance genes enrich the toolkit of plant pathology and may be represented in matrices that record the presence (blue square) or absence (grey square) of genes in the sequenced individuals. (f) Thanks to genome sequences, geneticists can include structural variants in their search for causal polymorphisms under GWAS peaks.