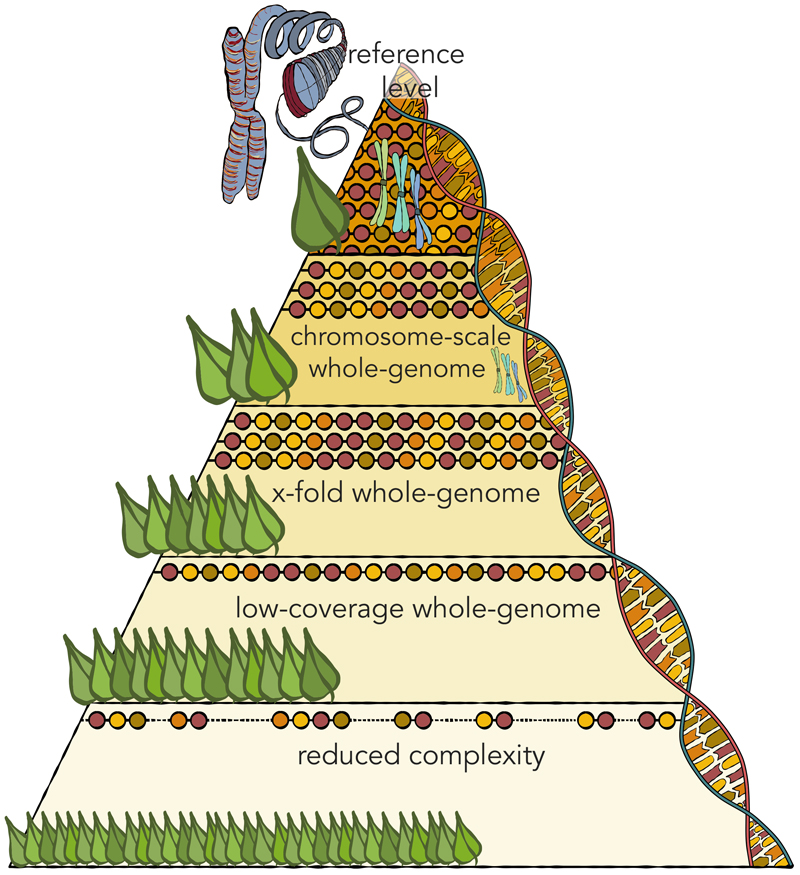
Figure 3. A tiered strategy for pangenomics.
Different sequence strategies (level of the pyramid) are suitable for different panel size (represented by leaf numbers). Reduced representation sequencing is done on as many genotypes, sampled in situ or from genebank collection, as possible. Representative coresets, sequenced to ever greater depth, are selected for different applications. Low-coverage (1- to 5-fold coverage) short-read whole genome sequencing aided by imputation is useful for genome-wide association scans and for genotyping known SVs. High-coverage (> 10-fold for inbred, > 30-fold for heterozygous genomes) short-read sequencing underpins selection scans, haplotype definition and demographic analyses. Genome assemblies based on long-read sequencing and chromosome-scale mapping catalogue the full spectrum of structural variation. Potentially extraordinary effort will be expended on a small number of genotypes to close gaps in difficult-to-assemble regions such as long tandem repeat arrays and centromeres to obtain telomere-to-telomere (T2T) assemblies. As technology progresses, the pyramid may turn into a cube and long-read sequencing may be employed in the bottom layers as well.