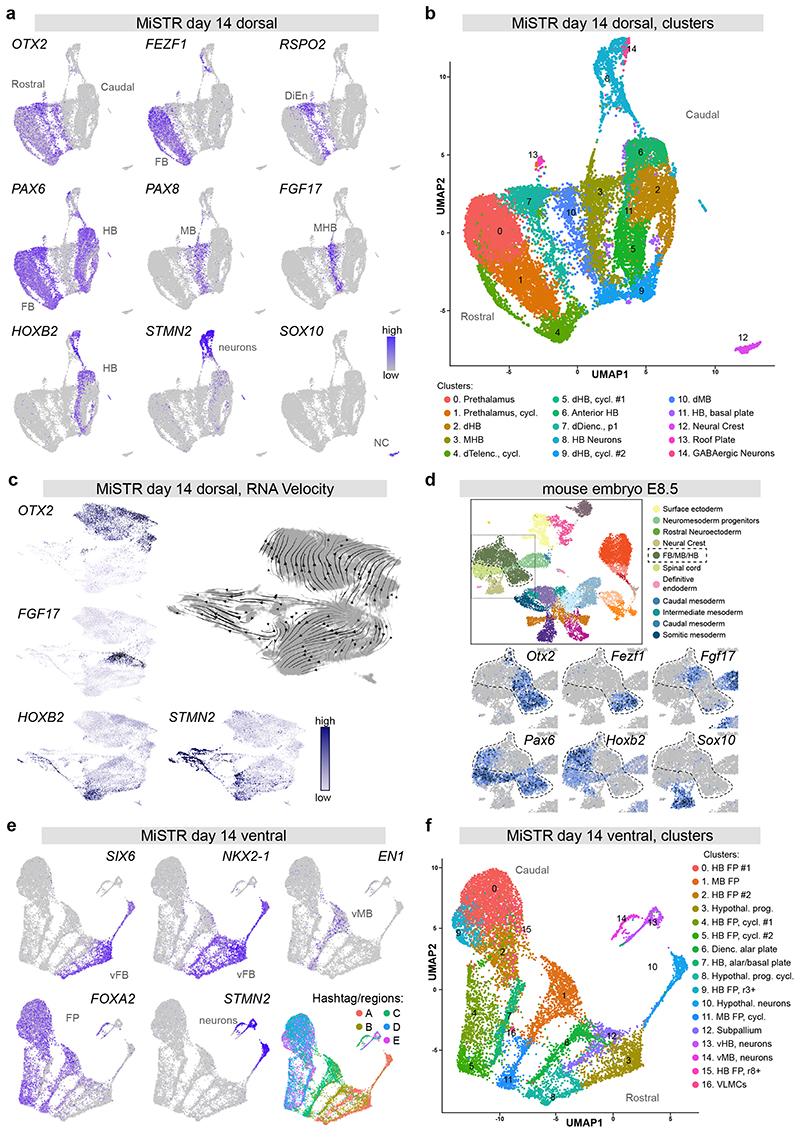
Figure 3. Single cell transcriptomics of dorsal and ventral MiSTR tissues.
(a, b) scRNAseq of 14 day dorsal MiSTR tissue (n=3 MiSTR tissues, 17683 cells) plotted in Uniform Manifold Approximation and Projection (UMAP) dimensions.
(a) Selected marker genes of neural patterning depict the rostro-caudal axis oriented left-to-right, revealed by the expression of genes marking the forebrain (FB), diencephalon (DiEn), midbrain (MB), midbrain-hindbrain boundary (MHB), and hindbrain (HB). Note the separate cluster of SOX10+ neural crest cells (NC) and a branch of STMN2+ neurons.
(b) Shared nearest neighbour (SNN) - based clusters calculated using Seurat. Cells are coloured according to their respective cluster and annotated based on literature and database search. dHB: dorsal Hindbrain, MHB: Midbrain-Hindbrain boundary, dTelenc: dorsal Telencephalon, cycl.: cycling, dDienc: dorsal Diencephalon, dMB: dorsal Midbrain. See also Supplementary Fig. 4a.
(c) RNA velocity analysis of day 14 dorsal MiSTR scRNAseq data (n=3 MiSTR tissues, 18241 cells), showing RNA trajectories away from the FGF17+ MHB cell cluster as well as towards postmitotic neurons (STMN2+), analysis performed in ScanPy.
(d) scRNAseq of E8.5 mouse embryonic stage plotted in UMAP dimensions showing selected marker genes of neural patterning, within the selected neural tube region (FB/MB/HB) highlighted in the overall map (top). See also Supplementary Fig. 4c. Plots were obtained from the scRNAseq map of mouse early development17, available at: https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018.
(e, f) scRNAseq of 14 day ventral MiSTR tissue (n = 3 MiSTR tissues, 12669 cells) plotted in UMAP dimensions.
(e) Expression of genes marking the ventral forebrain (vFB), ventral midbrain (vMB) and floor plate (FP) revealed a rostro-caudal axis oriented right-to-left in the UMAP. This is in accordance with the distribution of hashtag barcodes marking each of the MiSTR regions (A-E). See also Supplementary Fig. 6a for additional gene expression patterns.
(f) SNN- based clusters calculated using Seurat. Cells are coloured according to their respective cluster and annotated based on literature search. HB FP: Hindbrain Floor Plate, MB FP: Midbrain Floor Plate, Hypothal. Prog.: Hypothalamic progenitors, cycl.: cycling, Dienc.: Diencephalic, vHB: ventral Hindbrain, vMB: ventral Midbrain, VLMCs: Vascular leptomeningeal Cells.