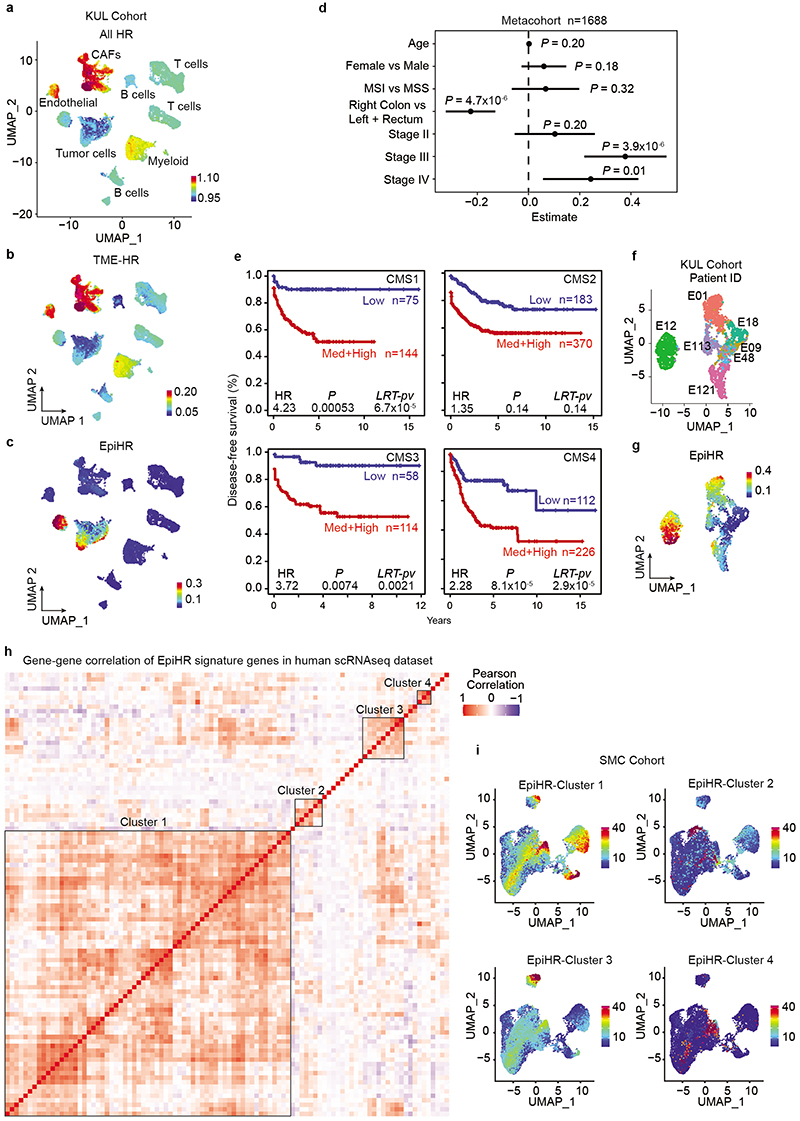
Extended Data Fig. 1 |. The EpiHR geneset marks a defined tumor cell population across CRCs.
a-c, UMAP layout of whole tumors (stroma + epithelium cells) from 7 CRC patients in the KUL dataset. Colored by (a) gene expression of all high hazard ratio genes (AllHR), (b) tumor microenvironment-specific HR genes (TME-HR), and (c) epithelial-specific HR genes (EpiHR). d, Association between clinical variables and the EpiHR signature in the CRC meta cohort was assessed by fitting a linear model for each variable independently. Technical factors (dataset and center, as described in extended methods) were included as covariates. Lines show the left and right confidence intervals. n= 1688 patients. e. Kaplan-Meier survival curves indicating relapse-free survival according to EpiHR gene signature expression for CRC patients classified by CMS. Two-sided Wald test. f-g, UMAP layout of 2718 CRC tumor cells from the KUL cohort colored by f) patient ID and g) expression of the EpiHR signature. h, Heatmap showing Pearson correlation scores in gene expression among EpiHR signature genes in patients from the SMC cohort. Note that most genes belong to one coherent subset (Cluster 1). Gene lists are detailed in Supplementary Table 2. i, UMAP layout of human CRC tumor cells colored by the expression of genes belonging to Clusters 1, 2, 3 and 4 identified in (h).