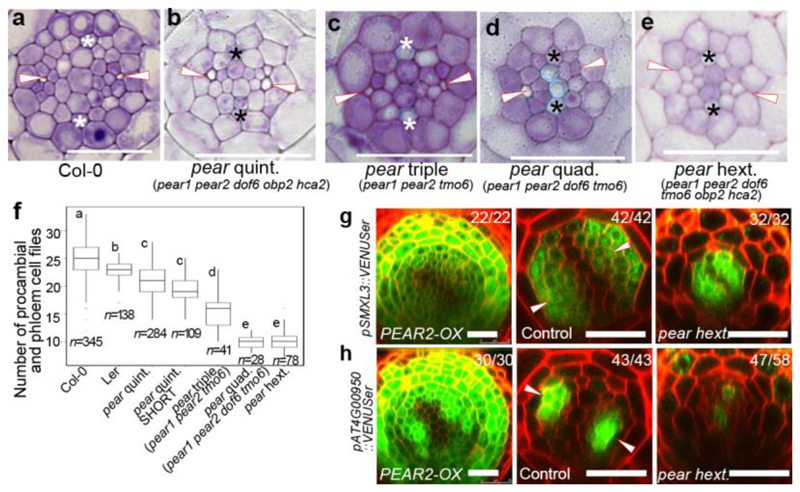
Figure 2. PEAR genes activate periclinal cell division by controlling downstream genes in non-cell autonomous manner.
a-e, Cross-section of wild type (a), pear quintuple (pear1 pear2 dof6 obp2 hca2) (b), pear triple (pear1 pear2 tmo6) (c), pear quadruple (pear1 pear2 dof6 tmo6) (d) and pear hextuple (pear1 pear2 dof6 obp2 hca2 tmo6) (e), respectively. Each image is representative of independent biological samples analysed in f. f, Number of procambial and phloem cell files in wild type and pear combinatorial mutants. Values were calculated from root cross sections at the differentiation zone. Boxplot centres show median. For more information on boxplots, see Methods. Statistically significant differences between groups were tested using Tukey’s HSD test p<0.05. For individual P values, see Supplementary Table 3. n, independent biological samples. g-h, Expression of selected PEAR1/2 downstream genes in wild type, PEAR2 overexpression plant and pear hextuple mutant. SMXL3 is expressed in phloem and procambial tissue, whose expression is induced by PEAR2 overexpression, but not altered in pear hextuple. AT4G00950 gene is expressed in PSE and its neighbouring cells, whose expression is induced by PEAR2 overexpression and reduced in pear hextuple mutant. Number in each panel indicates samples with similar results of the total independent biological samples analysed. White arrowheads, PSE. Asterisks, protoxylem. Scale bars, 25 μm.