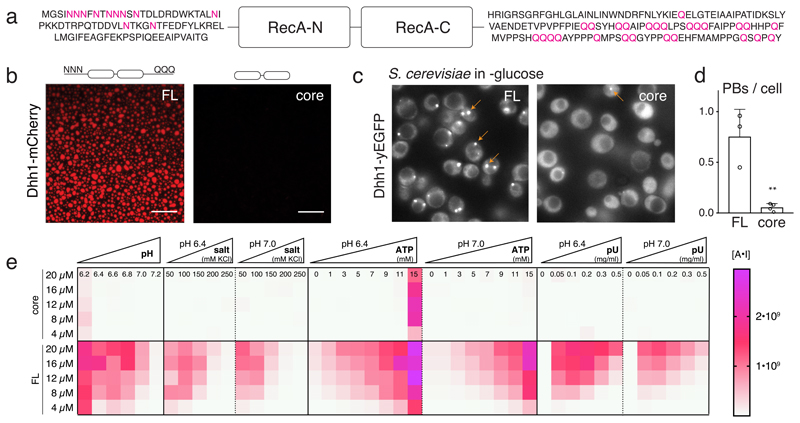
Fig. 1. The RNA-binding core and the unstructured tails of Dhh1 are required for LLPS and PB formation.
(A) Domain organization of Dhh1: RecA core and LCD tails. (B) In vitro phase separation of recombinant Dhh1-mCherry variants in the presence of ATP and RNA. Full-length (FL) and a truncation construct lacking both tails (core) were imaged at 10.5 μM protein. Representative images of at least 3 independent experiments. Scale bars: 25 μm. (C) Images of yeast cells expressing Dhh1-yEGFPs variants after 30 min glucose starvation. PBs are marked with arrows. Scale bars 5 μm. (D) Quantification of the number of PBs per cell using Diatrack. 3 (FL) or 4 (core) biological replicates of at least 417 (FL) or 505 (core) cells. Mean and SD; unpaired t-test (two-tailed), ** p-value = 0.0032. Dots represent the mean of individual replicates. (E) Phase separation behavior of full-length (FL) Dhh1 and the RecA core in response to changes in pH, salt, ATP and polyU concentration. Sum of the mean fluorescence intensity * area (arbitrary unit [A*I]) for all droplets per field of view, mean of 9 images (pH: 4 images).