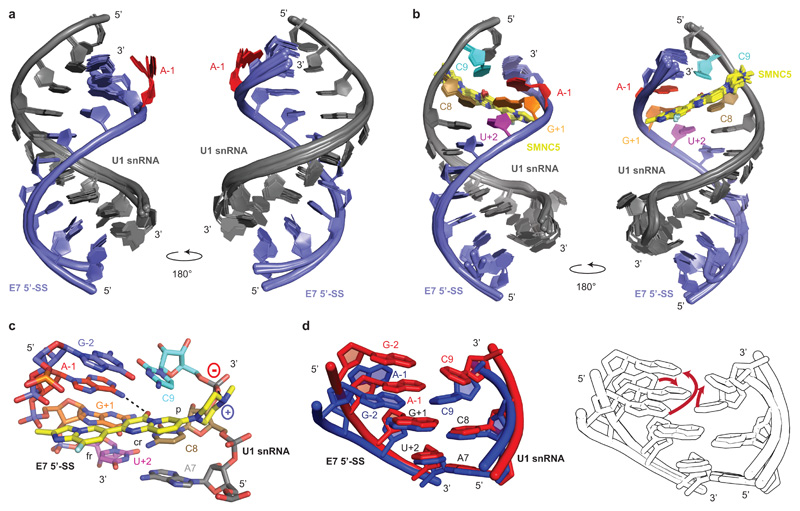
Fig. 2. Structural basis for SMN-C5 selectivity.
a, Overlay of the 20 NMR structures of the RNA duplex. The U1 snRNA and the E7 5’-SS are shown in grey and blue, respectively. The unpaired A-1 is highlighted in red. b, Overlay of the 20 NMR structures of the complex formed by the RNA duplex and SMN-C5. The U1 snRNA, the E7 5’-SS and SMN-C5 are shown in grey, blue and yellow, respectively. c, Closed up view of the SMN-C5 binding pocket. Black dashed lines illustrate the direct hydrogen bonds between SMN-C5 and the RNA. d, On the left, superimposition of the structures of the SMN-C5 binding pockets in the presence (red) or in the absence (blue) of SMN-C5. On the right, the same structures are represented colorless and red arrows indicate the RNA conformational change induced by SMN-C5.