Figure 2.
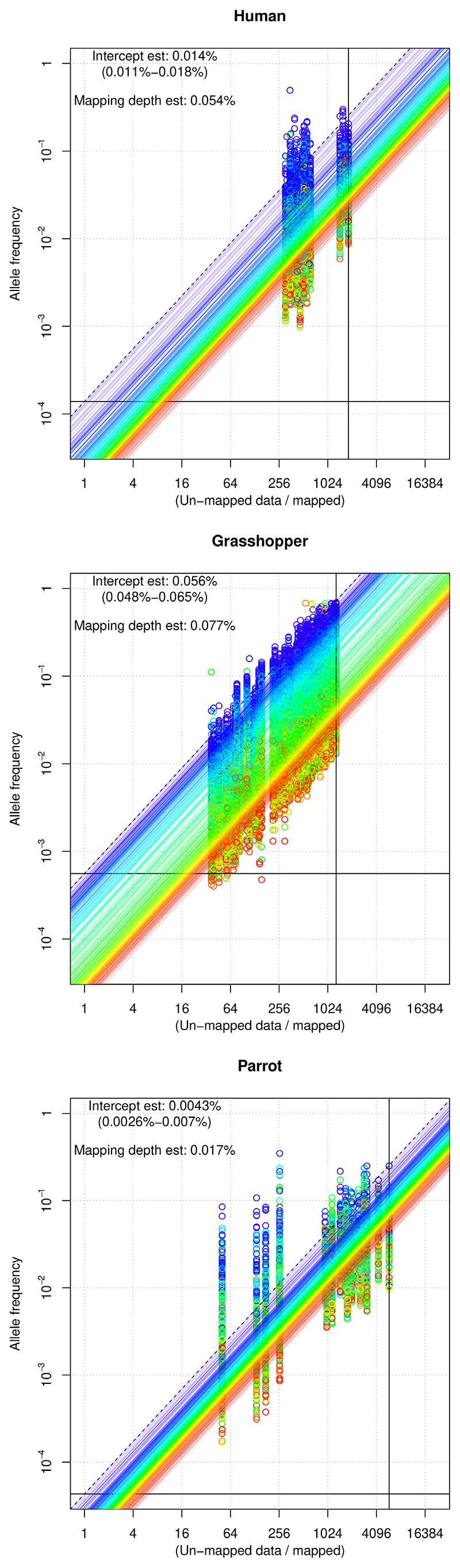
Change in the frequency of NUMT alleles with the mitochondrial mapping ratio (unmapped/mapped). The raw data are plotted as circles. The vertical stacks of points represent frequencies of different loci from the same sample, each locus being given a different colour (from red to violet according to the global average allele frequency). The lines with the corresponding colour show the best fit to Equation 1. Both axes are logarithmic. The intercept on the log scale will correspond to x = 1 (half the reads mapping to the mitochondrial genome, log(1) = 0). The two solid lines correspond to the minimum proportion of reads mapping to the mitochondrial genome across all samples (vertical) and to the intercept of the SNPs with the highest fitted allele frequency (horizontal). The dashed line shows the fitted relationship for the locus with the highest estimated allele frequency. The values of the horizontal solid and dashed lines at x = 1 are both estimates of the proportion of the genome composed of NUMTs (intercept estimate).
