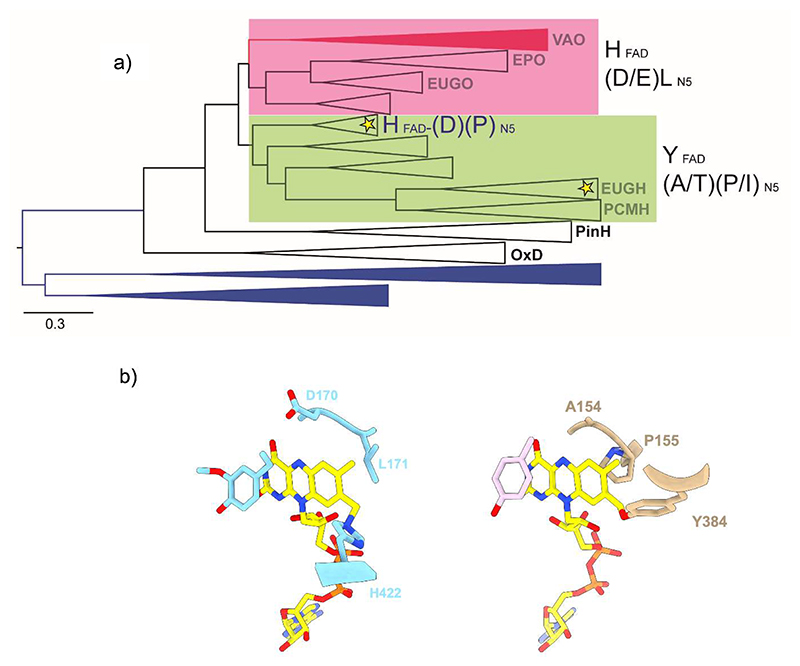
Figure 1. The VAO/PCMH enzymes acting on 4-phenol substrates.
(a) Condensed phylogeny. Color of branches indicate the taxonomic distribution: Archaea (blue), Bacteria (black) and Eukarya (red). The clades are names after their experimentally characterized, archetypal enzymes. OxD and PinH identify oxidative decarboxylases and pinoresinol hydroxylases, respectively; they are not part of this study. Oxidases are highlighted in pink, dehydrogenases in green. The locations of the enzymes identified in this work, vanillyl-alcohol dehydrogenase from Marinicaulis flavus and vanillyl-alcohol oxidase from Novosphingobium sp., are marked by yellow stars. On the right, the identity of the signature residues involved in the covalent binding or in contact with the N5 site of FAD are shown (Table 1). Please refer to Figure S1 for the fully annotated tree. The scale bar indicates substitutions per site. (b) Active site residues in VAO from Penicillium simplicissimum (right; PDB:2VAO) and PCMH from Pseudomonas putida (left, PDB:1DIQ).