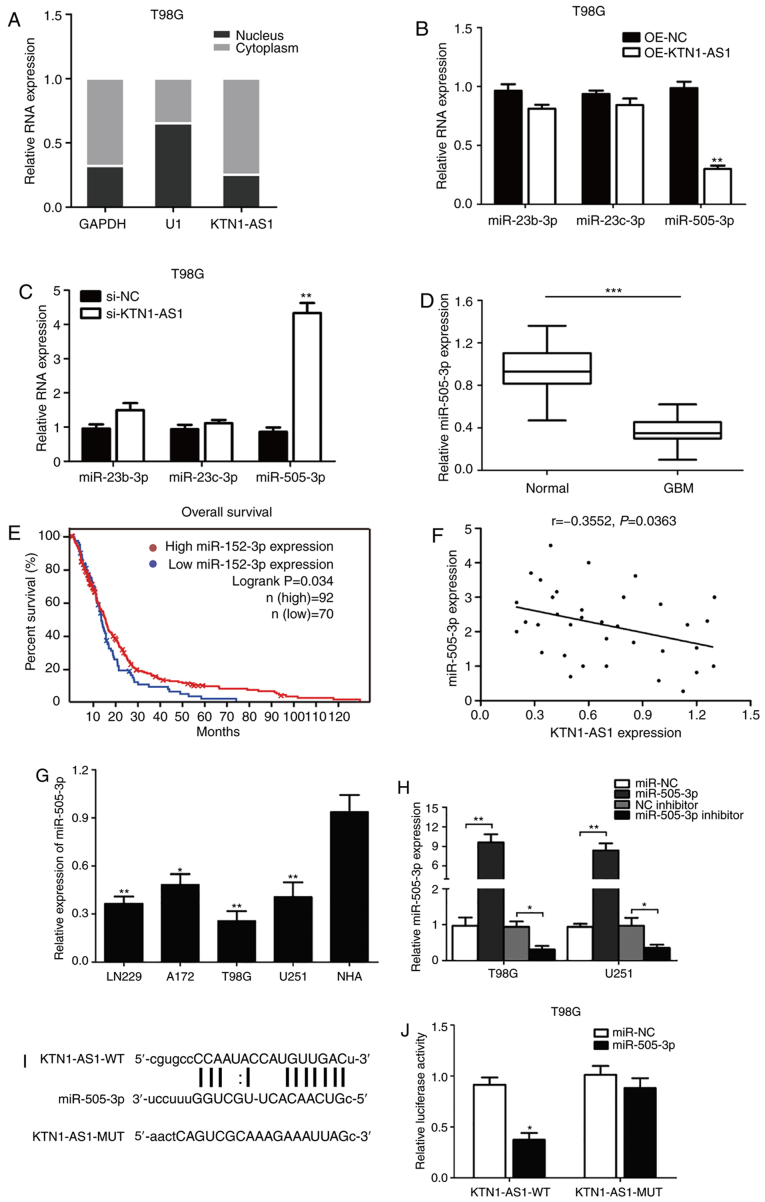
Figure 4.
KTN1-AS1 was targeted by miR-505-3p at 3′-UTR. (A) Expression of KTN1-AS1 in subcellular fractions was examined by RT-qPCR in T98G cells. Black range indicates nuclear fraction and gray range indicates cytoplasmic fraction. (B and C) Expression of the predicted binding miRNAs (miR-23b-3p, miR-23c-3p and miR-505-3p) was measured by RT-qPCR following transfection with si-KTN1-AS1 or OE-KTN1-AS1. (D) Decreased expression of miR-505-3p was identified in glioma tissues compared with normal tissues (P<0.001). (E) Patients with low level of miR-505-3p had better prognosis compared with patients with high miR-505-3p level in TCGA database (log-rank test; P=0.034). (F) Correlation between KTN1-AS1 and miR-505-3p levels was detected. (G) miR-505-3p level in glioma cell lines was analyzed by RT-qPCR. (H) miR-505-3p level was determined in T98G and U251 cells transfected with miRNA mimics/miR-NC or inhibitor/NC inhibitor by RT-qPCR. (I) Predicting binding sites of miR-505-3p on KTN1-AS1. Relative luciferase activity was measured after co-transfection with miR-505-3p mimics and psiCHECK2-KTN1-AS1-WT in T98G cells. (J) Luciferase activity was determined 24 h after transfection using a dual luciferase assay. Data were presented as the means ± standard error of the mean of at least three independent experiments. *P<0.05, **P<0.01 and ***P<0.001. KTN1-AS1, Kinectin 1 Antisense RNA 1; miR, microRNA; UTR, untranslated region; GBM, glioblastoma; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; si, small interfering; WT, wild-type; MUT, mutant; OE, overexpressing.