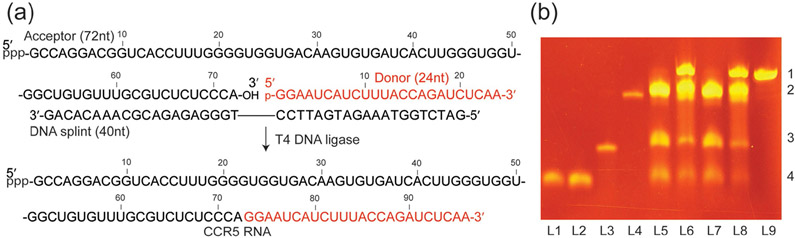
Figure 1.
Ligation of the CCR5 pseudoknot element. a) Scheme for preparation of segmentally and selectively 13C-labeled CCR5 RNA. The unlabeled 72 nt acceptor fragment was synthesized by in vitro transcription. The donor was prepared by either in vitro transcription followed by enzymatic dephosphorylation using RNA 5′ polyphosphatase or solid-phase synthesis followed by phosphorylation with T4 polynucleotide kinase to obtain the desired monophosphorylated group at the 5′-terminus. A 40 mer DNA splint was used to facilitate RNA ligation. b) A 12% denaturing PAGE showing the ligation results. L1: de-phosphorylated donor fragment obtained from in vitro transcription; L2: phosphorylated donor fragment from solid-phase synthesis; L3: DNA splint; L4: acceptor fragment ; L5: ligation reaction at 0 h using the donor from L1; L6: after 3 h reaction of L5; L7: ligation reaction at 0 h using the donor from L2; L8: after 3 h reaction of L7; L9: CCR5 mRNA pseudoknot prepared by in vitro transcription. Similar to prior observations,[8f] addition of the DNA splint to the reaction system resulted in changes of the migration pattern of all components in the system. The numbers at the right side of the image (1, 2, 3, and 4) correspond to full-length CCR5, 72 nt acceptor, DNA splint, and 24 nt donor fragment, respectively. Typically, excess amounts of the acceptor fragment and the DNA splint were applied and the ligation yield was >60% based on the conversion of the donor fragment which, in most cases, was prepared with different isotope labels.