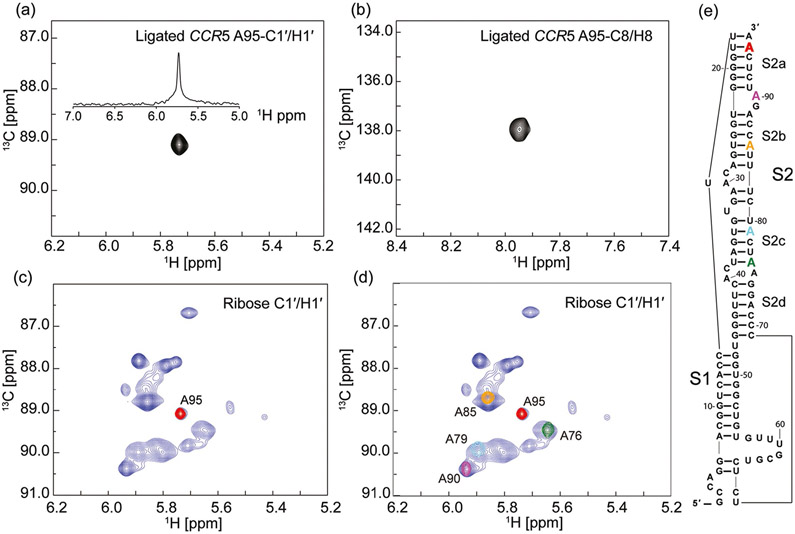
Figure 2.
Heteronuclear multiple quantum coherence (HMQC) spectra of the CCR5 pseudoknot element. a) Ribose C1′ and b) base C8 HMQC spectra of ligated CCR5 RNA bearing site-specific (8,1′)-13C-labeled probes at residue A95. The donor fragment was prepared by solid-phase synthesis on a DNA synthesizer first using unlabeled RNA amidites followed by incorporation of a selective (8,1′)-13C-labeled adenosine phosphoramidite at different positions. Upon ligation to the unlabeled acceptor fragment, the resulting ligated full length CCR5 pseudoknot element afforded a single peak at either ribose C1′ or base C8 region. The projected 1D 1H spectrum in part (a) displayed a single peak across a broader H1′ region, demonstrating the success in isotope labeling of a specific residue of the full length 96 nt RNA. c) Overlay of ribose HMQC spectra obtained from two CCR5 RNA pseudoknot samples. The spectrum in blue was recorded for a full length CCR5 RNA with all 17 adenosine residues labeled selectively with (8,1′)-13C atoms; the spectrum in red was from the ligated CCR5-A95 RNA. d) Ligation between the unlabeled acceptor and four different residue-specific labeled donor fragments resulted in resonance assignments of four additional residues: A90, A85, A79, and A76 at both ribose C1′ and base C8 regions. The spectrum in blue is the same as that in part (c). This method allowed for straightforward resonance assignments and provided staring points for assignment of the neighboring residues based on 2D/3D NOESY experiments. e) Secondary structure of CCR5 pseudoknot RNA based on previous SHAPE experiments.[4]