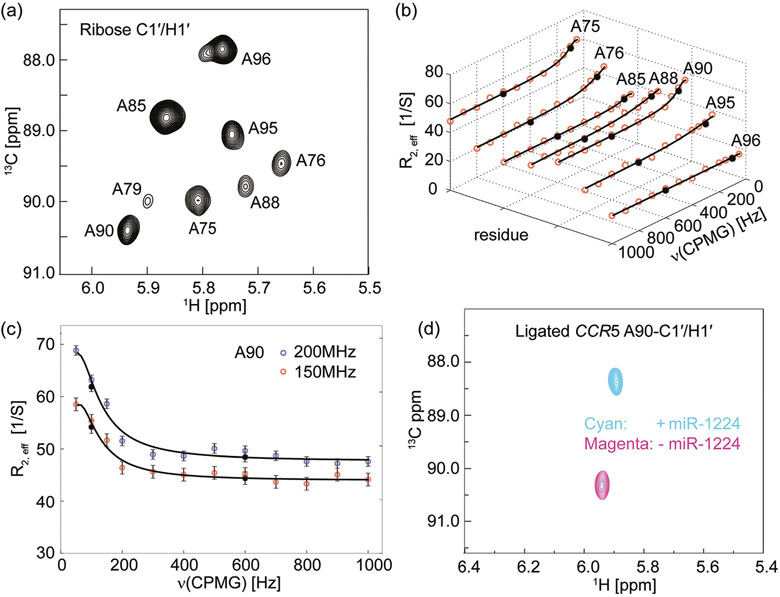
Figure 3.
Chemical exchange in CCR5 revealed by 13C Carr–Purcell–Meiboom–Gill relaxation dispersion (CPMG RD) experiments. a) 2D 1H–13C HMQC spectrum of a ligated CCR5 RNA sample labeled only with site-specific (8,1′-13C)-adenosines within the donor fragment. This was recorded in the absence of miR-1224, at 298 K, at 600 MHz proton Larmor frequency, with Trelax=0 ms. The expected eight anomeric H-C cross-peaks are well-resolved. b) 13C CPMG RD profiles of the ligated CCR5 RNA at 150 MHz 13C Larmor frequency. Red dots represent experimental data and black dots represent repeated experiments. Solid lines are best-RD-curve fits of the CPMG profiles. Residues A75, A76, A88, A90, and A95 show non-flat dispersion profiles whereas A85 and A96 show flat dispersion profiles. Note that the A79 resonance was too broad to adequately fit. c) 13C CPMG RD profiles of A90 ribose anomeric carbon atom of the ligated CCR5 RNA at 150 (red dots) and 200 MHz (blue dots) 13C Larmor frequency. Black circles represent repeated experiments and solid lines are the best fits of the CPMG profiles. d) NMR titration of the ligated CCR5-A90 RNA with miR-1224. The 2D HMQC spectrum in magenta recorded in the absence of miR-1224 exhibits a single cross-peak corresponding to ribose C1′ of the residue A90. Upon addition of miR-1224, the ribose A90-C1′ chemical shift changed dramatically from 90.3 ppm to 88.3 ppm (in cyan). The large chemical shift change (up to 2.0 ppm) indicates that the interaction with miR-1224 alters the local chemical environment of the A90 residues. In the presence of an equimolar amount of miR-1224, the CCR5 RNA is not completely converted to its bound form, with ca. 9% remaining in the free conformation. This is consistent with the EMSA result (Figure S6). However, adding 20% extra amount of miR-1224 converts it into the completely bound form (data not shown).