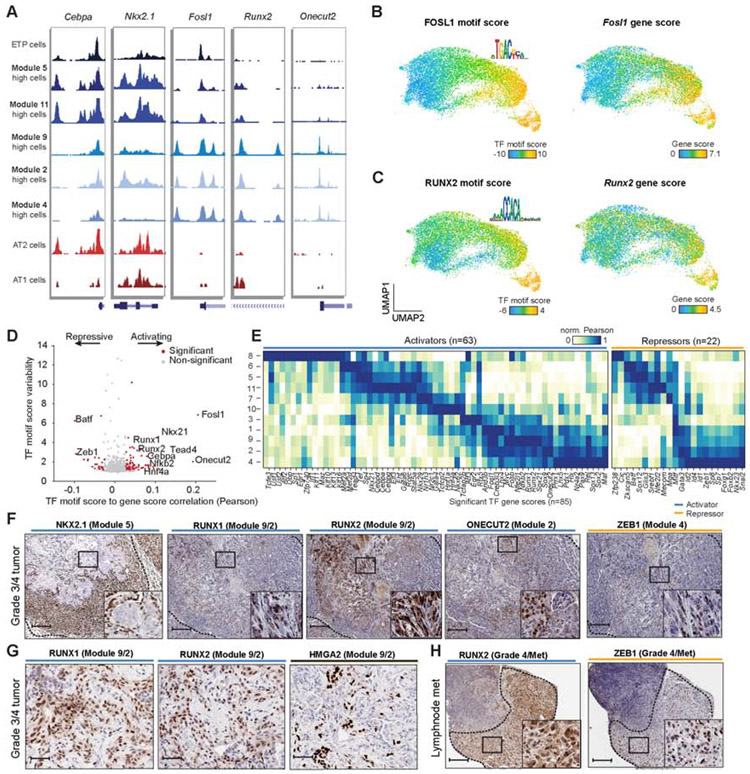
Figure 5. Regulatory analysis of cancer cells identified chromatin activators and repressors.
(A) Chromatin accessibility tracks for cells with high module scores and normal AT1/AT2 cells respectively at key transcription factors. Modules include early time point (ETP), early-stage (5, 11) and late-stage (9, 2, 4) modules. Module high was defined as two standard deviations above the mean module score across cells. (B-C) UMAP highlighting single-cell TF motif scores and motif logos (left), and gene scores (right) for FOSL1 (B) and RUNX2 (C) in cancer cells. (D) Correlation of TF motif scores with gene scores for each TF (n = 769) plotted against the TF motif score variability. Significantly variable TF motifs (motif score s.d. ≥ 1.2) correlated with their gene score (permutation p < 0.001) are shown in red; TFs with positive or negative correlation are highlighted as activators or repressors, respectively. Permutation p values were calcuated using a Z-test between the observed TF-motif gene correlation coefficient to the permuted correlation coefficients. TF motif scores signficiance was computed with deviation Z-scores across cells. (E) Normalized correlation (max/min normalized using Pearson r correlations) of TF gene scores to module scores delineated by activators (n = 58) and repressors (n = 14). (F) IHC of heterogeneous late-stage TFs stained for NKX2.1 (module 5), RUNX1 (module 9/2), RUNX2 (module 9/2), ONECUT2 (module 2), and ZEB1 (module 4) at 250 μm, inset 50 μm (n = 1). (G) Grade 4 regions stained for RUNX1, RUNX2 and HMGA2 (n = 1). (H) Lymph node tumors stain for RUNX2 and ZEB1 (250 μm, inset 50 μm; n = 1). see also Fig. S5 and Table S4.