Figure 1. Caspase Represses a Non-Stress p38 MAPK Function that Delays Post-Embryonic Development.
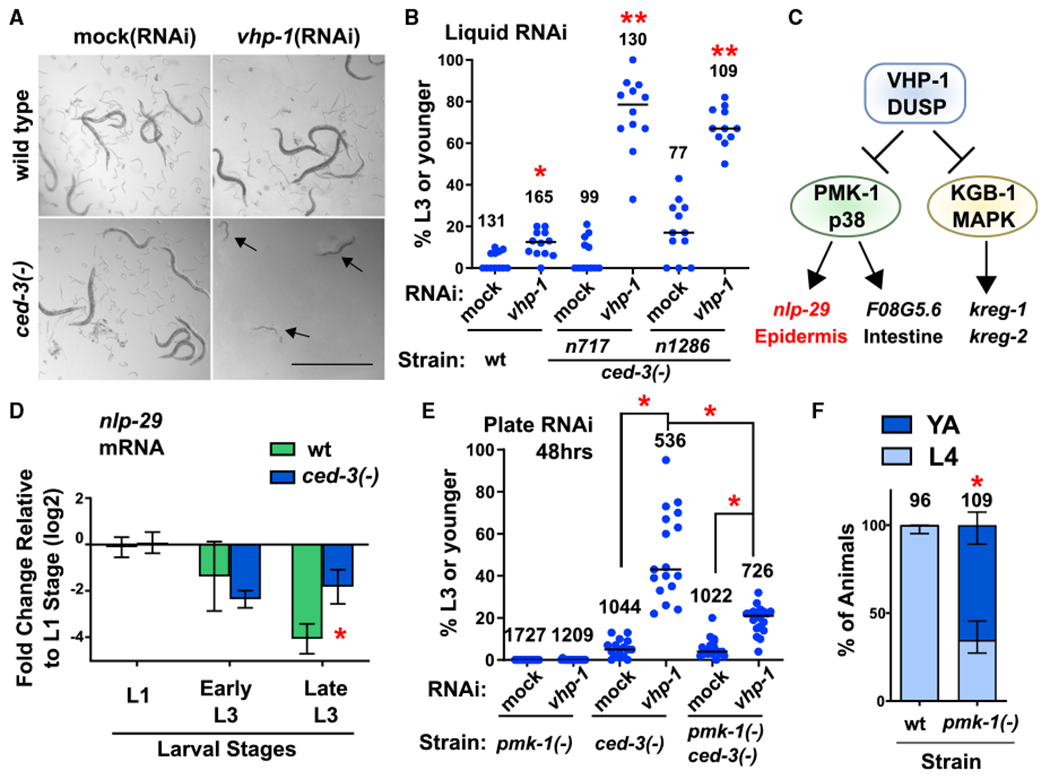
(A and B) Loss of both ced-3 and vhp-1 functions compromise viability. Images (A) and quantification (B) of worms in liquid RNAi cultures taken on a dissecting scope, showing a synergistic larval delay phenotype in ced-3(−) animals treated with vhp-1(RNAi). Magnification was the same throughout (scale bar indicates 1.0 mm). Arrows indicate P0 animals delayed in mid larval development. For all conditions, synchronous L1 stage P0 animals were added to the indicated food source (RNAi) and scored for developmental stage when negative control P0 animals had reached adulthood. Images in (A) show obvious presence of offspring (eggs and L1 stage larvae) in control wells. Quantified data are shown as dot plots indicating the percent of original P0 animals in the given well remaining at L3 stage or earlier, with total number of animals scored for each indicated condition. Data are pooled from three independent experiments and each dot corresponds to an individual well (four wells per experiment). Median values (horizontal bars). *Significant for wild-type treated with vhp-1(RNAi) compared to mock(RNAi), p = 0.0037, Mann-Whitney. **Significant for both ced-3(−) strains treated with vhp-1 (RNAi) compared to both mock(RNAi) of the given strain and wts train treated with vhp-1(RNAi), p < 0.0001, Mann-Whitney. For quantification of a ced-3(−) mutant generated by another lab, confirmation of ced-4(Apaf) requirement, and CED-3 caspase catalytic activity, see Figures S1A–S1C.
(C) The VHP-1 phosphatase is known to negatively regulate (black stops) PMK-1(p38 MAPK) and KGB-1 MAPK that activate (arrows) downstream genes (Kim et al., 2004; Richardson et al., 2010).
(D) qRT-PCR analysis demonstrates that CED-3 caspase limits the expression of epidermal PMK-1 downstream target nlp-29. *Significant for ced-3(−) strain compared to wt, p = 0.016, t test, standard deviation shown. Other PMK-1 and KGB-1 targets are not significantly upregulated by ced-3(−) mutation (Figure S1D).
(E) Loss of pmk-1 function helps restore larval development of ced-3(−) animals treated with vhp-1(RNAi) on agar plates. Data are shown as dot plots with total number of animals scored for each condition. Data are pooled from two independent experiments and each dot corresponds to an individual plate. Median values (thin horizontal bars). A significant restoration of development was observed with pmk-1(−) compared to animals with wild-type pmk-1 function. *Statistical comparisons indicated by brackets, p < 0.0001, Mann-Whitney.
(F) Effect of pmk-1(p38)(−) mutation alone on larval developmental rate. Wild-type and pmk-1(−) strains were assessed for the percent of animals at the fourth larval stage or adult stage 48 h after hatching. Proportion of animals for the given stage with standard deviation is shown, with total number of animals scored for each condition. *Significant, pmk-1(−) compared to wild-type, p < 0.0001, Fisher’s exact test. For confirmation of the PMK-1(p38) pathway and upstream factors in negatively repressing post-embryonic development in the absence of stress, see Figures S1E–S1G.
