Figure 6.
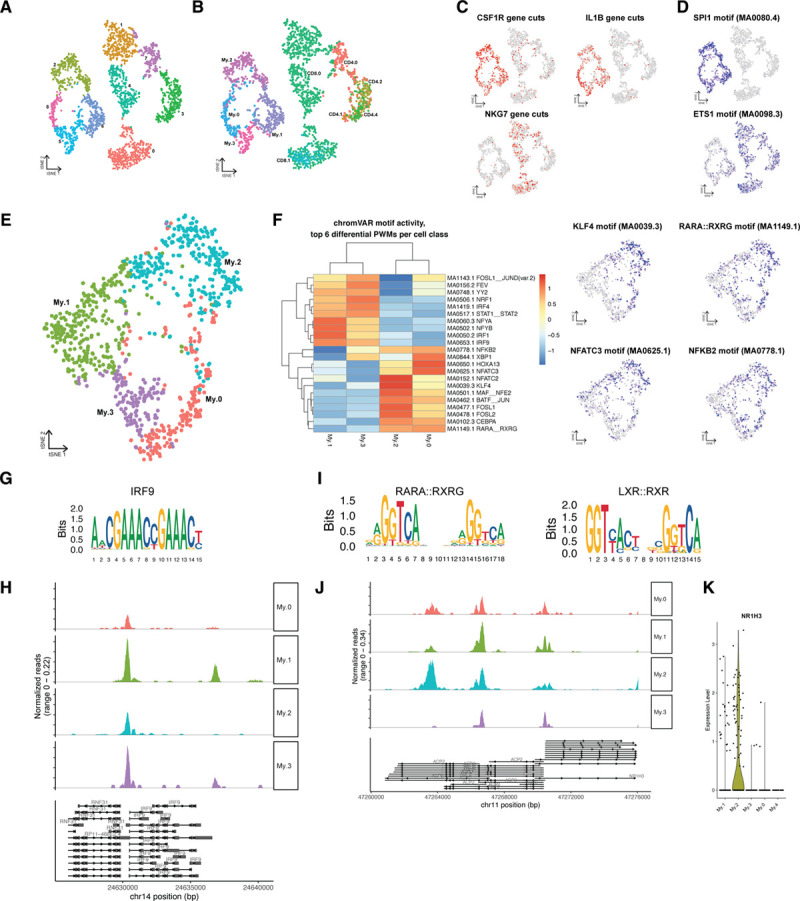
Chromatin accessibility of myeloid cells in human atherosclerotic plaques analyzed using single-cell ATAC sequencing (scATAC-seq). A, tSNE visualization of myeloid and T-cell clusters based on scATAC-seq. B, Projection of single-cell RNA sequencing (scRNA-seq) myeloid and T-cell labels over the scATAC-seq clusters. C, tSNE visualization of cell type–specific accessible gene loci. D, tSNE visualization of cell type–specific transcription factor motifs enriched in open chromatin regions. E, tSNE visualization of subclustered scATAC-seq myeloid clusters. F, Heatmap showing the top differential open chromatin TF motifs by chromVAR, with subcluster specific accessible TF motifs visualized as tSNE. G, IRF9 motif. H, Pseudobulk genome browser visualization identifying the open chromatin regions of IRF9 in different myeloid subsets. I, RARA:RXRG, and LXR (liver X receptor): RXR (retinoid X receptor) motifs. J, Pseudobulk genome browser visualization identifying open chromatin regions of NRIH3 (encoding LXRα) in different myeloid subsets. K, Violin plot of NR1H3 (nuclear receptor subfamily 1 group H) gene expression from myeloid scRNA-seq data. ETS1 indicates ETS proto-oncogene 1; IRF, interferon regulatory factor; KLF, kruppel like factor 4; NFATC3, nuclear factor of activated T cells 3; NR1H3, nuclear receptor subfamily 1 group H member 3; PWM, position weight matrix; tSNE, t-distributed stochastic neighbor embedding; RARA, retinoic acid receptor alpha; and RXRG, retinoid X receptor gamma.
