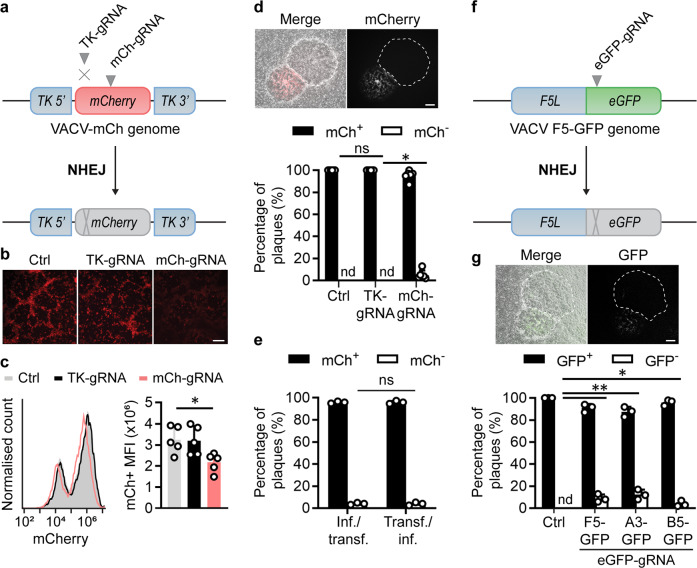
Fig. 1. Inefficient mutagenesis of VACV by CRISPR/Cas9-induced NHEJ.
a Schematic showing genome of VACV-mCh and the location of gRNA target sites. b, c Fluorescence micrographs (b) and flow cytometry (c) of VACV cultures 48 hrs after infection with VACV-mCh and transfection with the Cas9/gRNA complexes or no gRNA (Ctrl). Scale bar on micrographs = 200 μm (b); representative histogram and means and SEM of 5 replicates shown for MFI (*p = 0.0364 by one-way ANOVA) (c). d Progeny from cultures in (b) was grown on new monolayers and plaques scored for red fluorescence by microscopy. Example plaques are shown (scale bar = 300 μm). One hundred plaques from each culture across five independent experiments are graphed showing mean and SD (*p = 0.0037 by two-way ANOVA). e Experiment described above was recapitulated with reversed order of infection and transfection. f Schematic showing genome of VACV F5-GFP and gRNA target site as an example of the three additional viruses used. g Fluorescent plaque scoring was conducted as above (*p = 0.0150, **p = 0.0011 by two-way ANOVA, ns = not significant, nd = not detected, n = 3).