Figure 1.
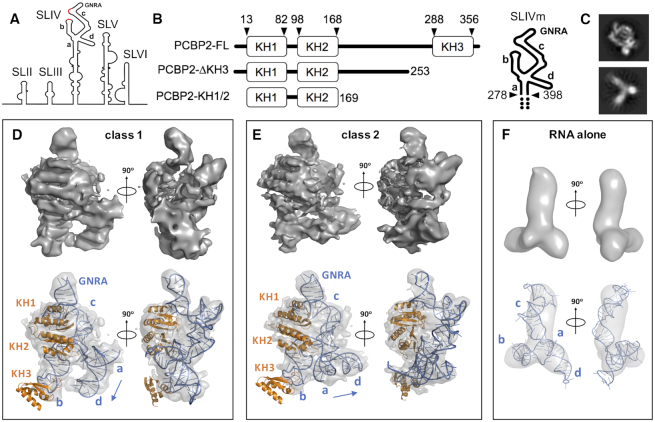
Cryo-electron microscopy analysis of PCBP2 interactions with SLIVm. (A) Schematic depicting the secondary structural arrangement of the poliovirus IRES. Highlighted are the C-rich regions (red) and GNRA tetraloop structure at the apex of SLIV. (B) Schematic representation of the protein and RNA constructs prepared for the current study. PCBP2 constructs included the full-length native sequence (PCBP2-FL), PCBP2 with KH3 removed from the 3CD cleavage site (PCBP2-ΔKH3) and a PCBP2 construct representing just the first two KH domains (PCBP2-KH1/2). The SLIVm RNA comprised bases 278–398 and three extra GC base pairs at the termini (depicted by dots). (C) Representative 2D classes for PCBP-FL/SLIVm (TOP) and SLIVm alone (bottom); (D, E) Top: Cryo-EM surfaces derived for class 1 and class 2 3D reconstructions of the PCBP2-FL/SLIVm complex. Bottom: constructed models of the PCBP2/SLIVm complex superposed with transparent cryo-EM surfaces to which the models were fitted. (F) Top: Cryo-EM surface derived for SLIVm alone from 3D reconstruction. Bottom: SLIVm model fitted to the cryo-EM surface, showing a similarity to the expected topology of the RNA.