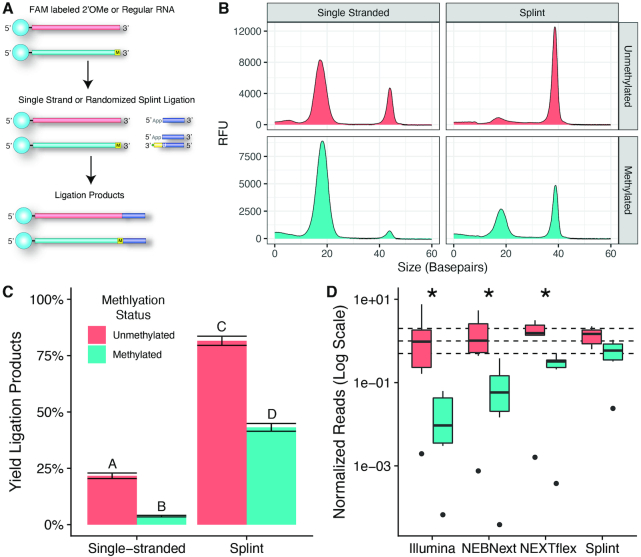
Figure 5.
Ligation and sequencing of 2′OMe modified RNA. (A) Schematic of the CE assay. A FAM labelled degenerate RNA oligo containing either a normal ribonucleotide or a 2′OMe modified at the 3′ terminus was used as a substrate for ligation reactions using single-stranded or randomized splint methods. (B) Example CE traces. Shorter unligated substrate runs faster at around 18 bp, while the longer ligation products run slower at >35 bp. (C) Percentage of the FAM labeled oligo that was converted to the ligation product. Values represent the mean of four replicates ± the standard deviation and letters represent groups that are significantly different from each other with a Tukey corrected P-value < 0.0001. (D) Normalized number of 2′OMe spike-in reads plotted on a logarithmic scale. Each miRNA was expected to have a normalized read value of 1 (central dashed line). The upper and lower dashed lines correspond to the interval of 2-fold over- or under-represented. Asterisks denote groups where the normalized read counts of the unmethylated oligos was significantly different from the methylated oligos at a Tukey corrected P-value of < 0.05.