Figure 3.

(A) Diagram of sequence-structure profile (SSP). SSP is computed for each unique sequence, and consists of a sequence profile (4 × unique sequence length) representing the nucleotide sequence and a secondary structure profile (6 × unique sequence length) representing the secondary structure. The sequence profile is a matrix computed by converting the nucleotide sequence of each unique sequence into a one-hot vector using a real value 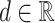 . The secondary structure profile is a matrix computed by converting the data obtained from CapR. (B) An example image of RNA secondary structure. The dashed lines represent base pairs. The secondary structure features predicted by CapR are classified into six types (bulge, exterior, hairpin, internal, multibranch, stem).
. The secondary structure profile is a matrix computed by converting the data obtained from CapR. (B) An example image of RNA secondary structure. The dashed lines represent base pairs. The secondary structure features predicted by CapR are classified into six types (bulge, exterior, hairpin, internal, multibranch, stem).
