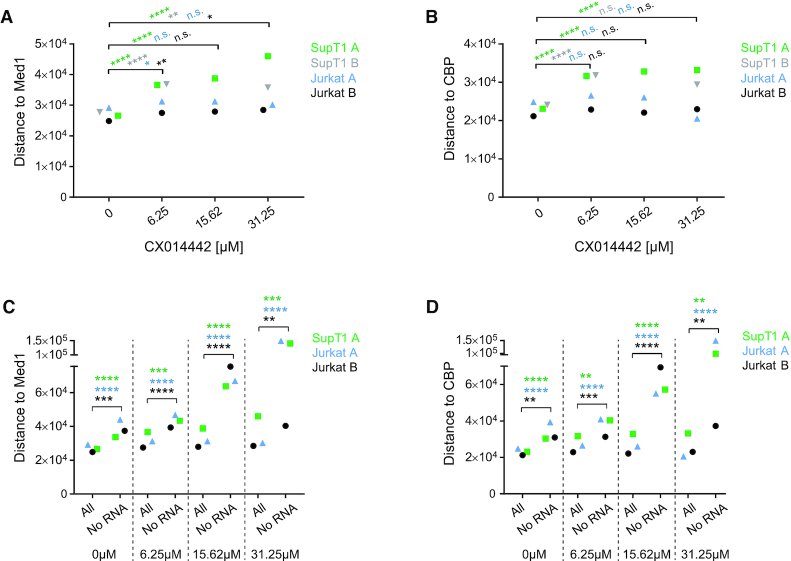
Figure 6.
Effect of LEDGINs on super-enhancer markers Med1 and CBP. (A, B) Integration site analysis in SupT1 and Jurkat cells determined the distance in base pairs of HIV provirus to super-enhancer markers Med1 (A) and CBP (B) in each condition (0, 6.25, 15.62 or 31.25 μM of CX014442). The median distance (bp) is plotted on the y-axis for two independent experiments in Jurkat cells (experiment A blue, experiment B black) and two in SupT1 cells (experiment A green, experiment B gray). (C, D) The distance in base pairs (bp) of integration sites to Med1 (C) and CBP (D) was determined for ‘no RNA’ sites in each condition (0, 6.25, 15.62 and 31.25 μM of CX014442) and plotted next to ‘all sites’. The median distance (bp) is plotted on the y-axis for two independent experiments in Jurkat cells and one experiment in SupT1 cells. Statistical significance was calculated by the Kruskal–Wallis test, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001. bp; base pairs, Med1; Mediator 1, CBP; CREB-binding protein.