Figure 1.
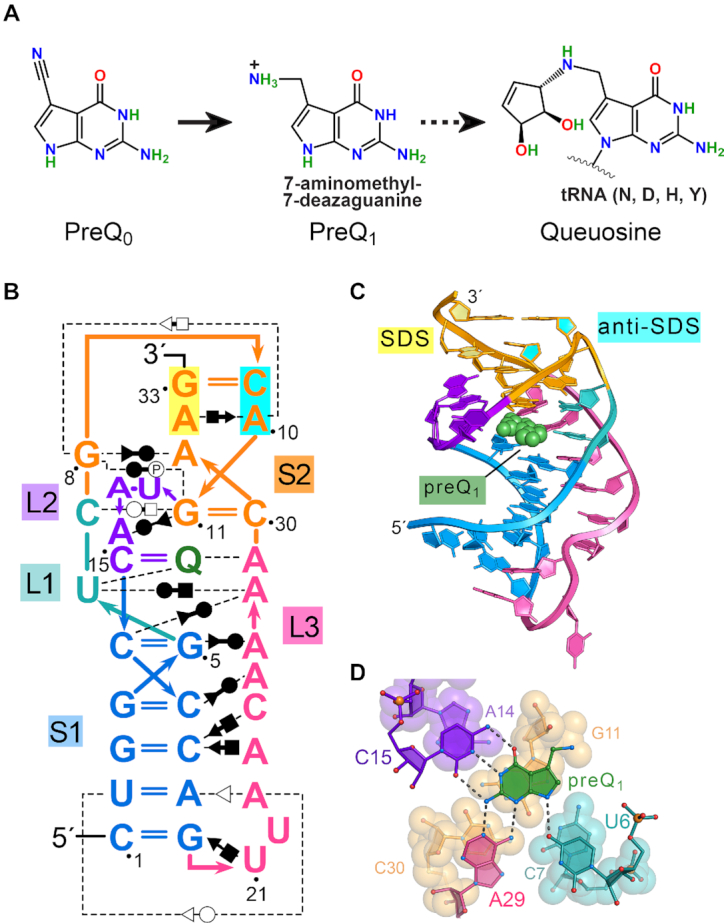
Queuosine (Q) synthesis, the Tte preQ1-I riboswitch fold and mode of effector binding. (A) The hypermodified base Q is synthesized by many bacteria starting from GTP and proceeds via pyrrolopyrimidine intermediates prequeuosine0 and prequeuosine1 (preQ1). PreQ1 is the last soluble metabolite prior to insertion into GUN anticodons of tRNAs decoding Asn, Asp, His or Tyr (broken arrow). In situ modifications are then added to the nucleotide to form Q (93). (B) Secondary structure of the wild-type preQ1-I riboswitch from Thermoanaerobacter tengcongensis (Tte) based on prior co-crystal structures (22,35). Color-codes correspond to specific pseudoknot stem (S) and loop (L) sequence as defined (94); preQ1 is shown as ‘Q’ (green). Non-canonical base pairing is indicated by the Leontis–Westhof symbols (95). Interactions with phosphate groups are denoted with the symbol P. The Shine-Dalgarno sequence (SDS) (expression platform) is highlighted in yellow; the opposing anti-(a)SDS is highlighted in cyan. (C) Ribbon diagram depicting the H-type pseudoknot fold based on a prior Tte co-crystal structure (PDB entry: 3q50) (22). Helix S2 comprises SDS-aSDS pairing (base and ribose rings filled yellow and cyan) that supports the gene-off conformation of the expression platform. PreQ1 is depicted as a CPK model (green). (D) Close-up view of the preQ1 binding pocket rotated −90° about the x-axis from panel C. Nucleotides that contribute to preQ1 specificity are depicted as ball-and-stick models; specificity base C15 of loop L2 forms Watson-Crick-like pairing to the effector face. Effector-recognition bases U6 and A29 contribute hydrogen bonds to the minor-groove (equivalent) edge of the effector. Here and elsewhere, putative hydrogen bonds are depicted as dashed lines. Nucleotides beneath preQ1 are depicted as CPK models.
