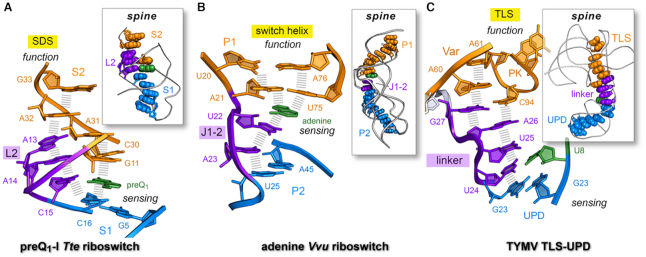
Figure 8.
Long-range nucleobase interaction spines in riboswitches and the TYMV 3′-UTR. (A) Stacking spine of the preQ1-bound Tte riboswitch (PDB entry: 3q50) (22). The spine exhibits continuous nucleobase stacking (hash marks) supported by preQ1 binding at C15 in loop L2; the resulting stacking spine communicates effector binding in the sensing region (aptamer) to the distal SDS (expression platform) to support a gene-off functional state; U12 was omitted for clarity. Inset, preQ1 favors a continuous stacking spine (space-filling model) that spans the length of the riboswitch. (B) Stacking spine of the Vvu add riboswitch in complex with adenine (PDB entry 5swe) (3). Adenine mediates nucleobase stacking contacts between switch-helix P1 and J1–2 nucleobases (hash marks). Inset, a continuous stacking spine spans the length of the riboswitch and includes adenine, which stabilizes the fold to achieve gene-regulation. (C) Stacking spine of the TYMV TLS-linker-UPD domain interaction (PDB entry: 6mj0) (1). In the absence of ribosomes, the UPD sensing domain remains intact and favors continuous base stacking that extends from the UPD, through the linker and into the TLS (hash marks). The U8•U24 pair (U8 in green) is analogous to effector binding by a riboswitch, albeit several other UPD bases influence stability of this domain (12). Inset, a continuous stacking spine spans the length of the structure. In each example, the designation ‘spine’ is justified due to the global stability imparted by the motif to the entire fold; in this manner, the spine provides a tangible communication conduit that links a sensing region with a functional domain (1).