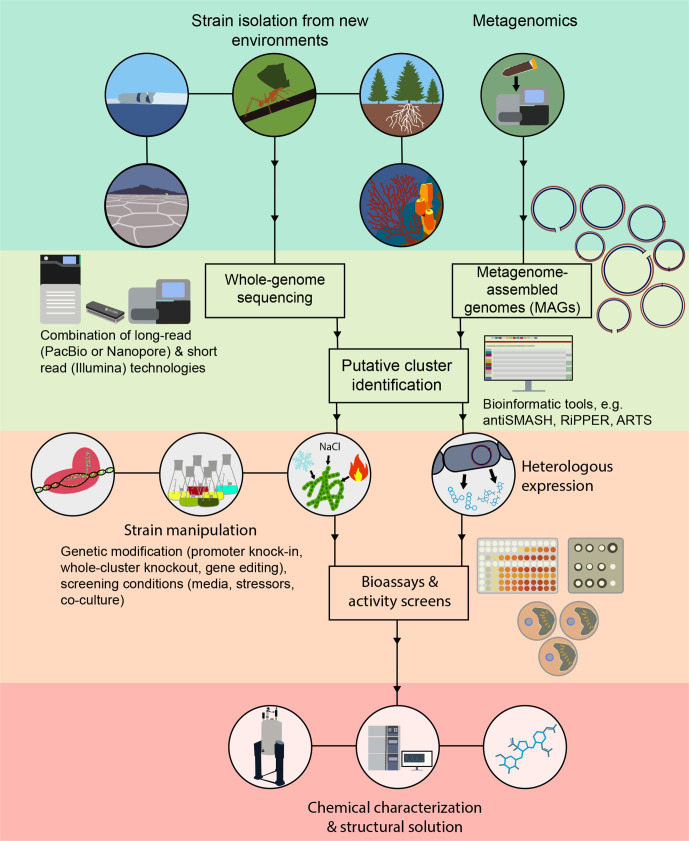
Fig. 2.
Graphic showing the general antibiotic compound discovery pipeline. The process usually begins with the isolation of novel strains or DNA from environmental samples. Once genomes and metagenomes are sequenced and assembled, the sequences can be analysed with tools such as RiPPER, antiSMASH or ARTS to identify biosynthetic gene clusters (BGCs). A combination of cluster synthesis, cloning and heterologous expression, or strain manipulation through CRISPR or varied conditions, can then be used to encourage the production of novel antibiotics. The production of antibiotics is screened for using bioactivity assays. Compounds can then be further characterized using techniques such as NMR or mass spectrometry.