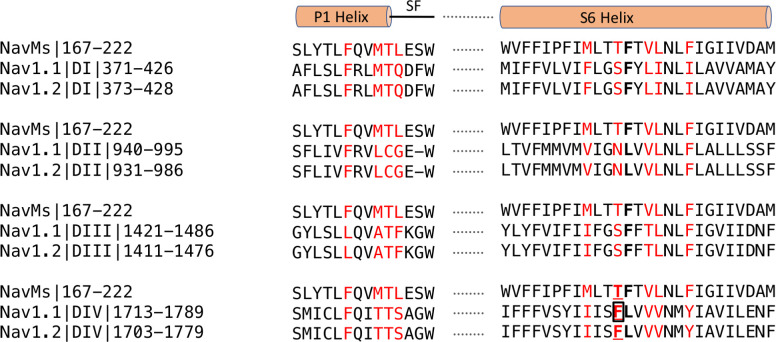
Figure 6. Sequence alignments of CBD-binding site regions of NavMs With corresponding regions in hNav1.1 and hNav1.2.
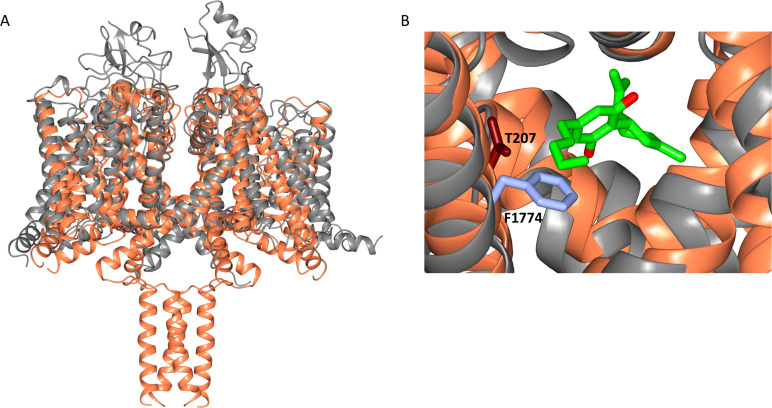
In red are the CBD-binding residues (within 3.9 Å of the compound) in NavMs and the equivalent residues in hNav1.1 and hNav1.2. The bold black F indicates the site of the NavMs F208L mutant used in these studies. It was changed from F to L in NavMsL because in half of the human Nav domains it is an F and in the other half it is an L (both indicated in bold black in the other sequences). However, as shown in Figure 6—figure supplement 2, the residue type present at this site makes essentially no difference in the structure. Residues located in the binding site are found within the P1 pore helix, the selectivity filter loop, and the S6 helix. The residue, which when mutated to alanine in hNav1.1 reduces the binding affinity of CBD (Ghovanloo et al., 2018), is boxed. This corresponds to T207 in NavMs, which is the residue that was mutated to alanine in the electrophysiology characterisations in the present study. The sequence alignment was carried out using Clustal Omega (Sievers et al., 2011) and annotated manually.